Professional Documents
Culture Documents
Lec 014 Gene To Protein
Uploaded by
QusizleOriginal Description:
Original Title
Copyright
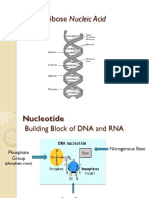
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Lec 014 Gene To Protein
Uploaded by
QusizleCopyright:
Available Formats
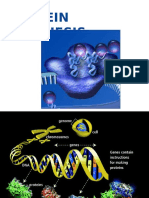
LECTURE #13 FROM GENE TO PROTEIN 1. FROM GENE TO PROTEIN 2. GENE EXPRESSION A.
. DNA inherited by an organism leads to specific traits by DICTATING THE SYNTHESIS OF PROTEINS and of RNA molecules involved in protein synthesis. B. Proteins are the link between genotype and phenotype. 3. ONE GENE ONE POLYPEPTIDE 4. GENES SPECIFY PROTEINS a. Alkaptonuria (black urine disease or alcaptonuria) is a rare inherited genetic disorder of tyrosine metabolism. This is an autosomal recessive condition that is due to a defect in the enzyme homogentisate 1,2-dioxygenase which participates in the degradation of tyrosine. As a result, a toxic tyrosine byproduct called homogentisic acid (or alkapton) accumulates in the blood, and is excreted in urine in large amounts(hence -uria). b. Excessive homogentisic acid causes damage to cartilage (ochronosis, leading to osteoarthritis) and heart valves as well as precipitating as kidney stones. 5. BEADLE & TATUM - Nobel Prize 1958 A. Beadle and Tatum identified mutant Neurospora crassa bread mold that could not survive on minimal medium because they were unable to synthesize certain essential molecules from minimal medium. b. All mutants could survive on a complete growth medium. Tested each individual mutant in a different vial that provided a single nutrient. c. The nutrient vial that allowed growth indicated the metabolic defect (thus, the gene with a mutation). d. Beadle and Tatum dubbed their hypothesis the one gene-one enzyme hypothesis. 6. OVERVIEW: Transcription and Translation a. Genes provide the instructions for making specific proteins, BUT DOES NOT BUILD THE PROTEIN DIRECTLY. b. The message is carried from DNA by RNA to the ribosome where synthesis can take place. 7. COMPARE DNA and RNA a. See chapter 5 for chart in textbook b. Remember, there are slight differences between DNA and RNA. c. RNA is chemically similar to DNA except that it contains: 1. Ribose as its sugar instead of deoxyribose. 2. The nitrogenous base uracil instead of thymine. 3. An RNA molecule is usually single stranded. d. Genes are typically hundreds to thousands of nucleotides long, each gene having a specific sequence of bases. e. Polypeptides are monomers long (think of the proteins primary structure, but the monomers are amino acids. f. How do we get from the language of nucleotides to the language of amino acids? 8. TRANSCRIPTION a. Transcription is the synthesis of RNA under the direction of DNA. b. Both molecules use the same language, so it is a simple copying of the information. Like a scribe would copy manuscripts before we had Xerox machines.
c. The copies are mRNA or messenger RNA. 9. TRANSLATION a. Translation is the synthesis of a polypeptide (the making of a protein) under the direction of an mRNA. b. The cell must translate the base sequence of an mRNA molecule into the language of amino acids. c. The site where the translation occurs is the ribosome. 10. PRO MEANS NO, EU MEANS TRUE a. Transcription and translation occur in the cytoplasm in prokaryotes because there is NO membrane bound nucleus. b. Transcription occurs in the nucleus and translation occurs in the cytoplasm of eukaryotes because there is a true membrane bound nucleus. 11. RICIN AND RIBOSOMES a. Ricin comes from the castor oil plant where it is concentrated in the seeds. b. A dose of ricin the size of a grain of salt can kill you. c. Ricin inactivates ribosomes. d. No ribosomes, no proteins, no life. 12. DNA RNA PROTEIN a. Genes program protein synthesis by messenger RNA. b. Francis Crick (of Watson and Crick) dubbed this the central dogma or the information flow in the cell. c. What language is used in translation? 13. THE GENETIC CODE a. How many bases correspond to an amino acid? b. Elaborate experimentation determined that triplets of nucleotide bases specify all amino acids. c. How do you read the code? d. In the language of mRNA. 14. DURING TRANSLATION a. mRNA base triplets are called CODONS. b. The codons are read in the 5 to 3 direction along the mRNA. c. Each codon specifies which one of the 20 amino acids will be added to the growing polypeptide chain. d. Codons are base triplets therefore; the nucleotides must be 3 times the number of amino acids. 15. GENETIC CODE PARTICULARS a. Complete code deciphered by mid-1960s. b. 61 of the triplets code for amino acids. c. Three codons that do not code for amino acids are stop signals or termination codons. They end translation. d. Codon AUG has two functions: to start an amino acid chain or to add a methionine. e. REDUNDANCY NO AMBIGUITY! 16. AUGTHEREDDOGATETHECATUAA a. READING FRAME IS KEY b. Spaces between the codons are unnecessary.
c. THE RIBOSOMES READ THE MESSAGE IN NON-OVERLAPPING THREE LETTER WORDS THAT ARE IN THE CORRECT ORDER. d. Incorrect ORDER: e. AUGTHEREDCATATETHEDOGUAA Not the intended message oops! f. AUGTHEREDGODTEAHECATUAA g. This is the stuff that DNA mutations are made of WITH SERIOUS CONSEQUENCES. 17. DNA POINT MUTATIONS A. Base pair substitutions: b. Silent a base pair substitution that has no effect on amino acid sequence. c. Missense A base pair substitution that results in a codon that codes for a different amino acid. d. Nonsense A mutation that changes an amino acid to one of the three stop codons resulting in a shorter (truncated) and usually nonfunctional protein. 18. DNA POINT MUTATIONS A. INSERTIONS OR DELETIONS: additions or losses of nucleotide pairs in a gene. B. Frameshift mutation: A mutation occurring when the number of nucleotides inserted or deleted is NOT A MULTIPLE OF 3, resulting in the improper grouping of the subsequent nucleotides into codons. C. Frameshift causing missense. D. Frameshift causing nonsense. E. 3 base-pair deletion or insertion, no frameshift, extra or missing amino acid. 19. EXPRESSING GENES FROM DIFFERENT SPECIES a. A NEW WAY OF LIGHTING UP! b. A tobacco plant expressing a firefly gene. c. How? The genetic code is nearly universal. d. Example: The RNA codon CCG is translated into amino acid proline in all organisms whose genetic code has been examined. 20. TRANSCRIPTION CLOSE UP a. Enzyme RNA polymerase opens the double-stranded DNA and joins the RNA nucleotides as they base-pair along the DNA template. b. Must assemble in the 5 3 direction. c. RNA polymerase does not need a primer. 21. TRANSCRIPTION CLOSE UP a. Specific sequences along the DNA mark where the RNA polymerase attaches and initiates transcription the PROMOTER. b. In bacteria, it ends at the TERMINATOR. c. The direction of transcription is downstream. The promoter is upstream from the terminator. 22. TRANSCRIPTION INITIATION AT A EUKARYOTIC PROMOTER a. Greater complexity in eukaryotes: a transcription initiation complex is required to start transcription. b. Eukaryotic promoter has a TATA box about 25 nucleotides upstream from start point. One of several transcription factors will recognize the TATA box and bind to the DNA before the RNA polymerase II can do so. c. DNA unwinds, transcription begins. 23. ELONGATION
a. RNA polymerase moves along the DNA, untwisting the double helix, exposing 10-20 DNA bases at a time to pair with RNA nucleotides. b. Nucleotides are added to the 3 end of the growing RNA molecule. c. RNA peels away as it grows, DNA double helix re-forms, at approximately 40 nucleotides per second in eukaryotes. d. Non-template strand often referred to as the coding strand note what it matches! 24. A SINGLE GENE CAN BE TRANSCRIBED SIMULTANEOUSLY BY MANY RNA POLYMERASES a. A growing strand of RNA trails off from each RNA polymerase along the DNA. b. Simultaneous transcription allows for many mRNA transcripts to increase production of the encoded protein. 25. TERMINATION a. Different mechanism in bacteria and eukaryotes. b. Bacteria: a terminator sequence in the DNA is transcribed and causes the polymerase to detach and release the transcript. c. Immediately available as is for use as mRNA. 26. TERMINATION - Eukaryotes a. AAUAAA: polyadenylation signal sequence. b. In eukaryotes, RNA polymerase II transcribes a sequence on the DNA called the polyadenylation signal sequence seen in pre-mRNA (the primary transcript before modification). c. 10 to 35 nucleotides downstream from the AAUAAA signal, proteins associated with the growing RNA transcript cut it free from the polymerase. d. The polymerase will continue transcribing for hundreds of nucleotides past the site where the mRNA was cut free currently being researched. 27. RNA PROCESSING a. After transcription, pre-mRNA must be processed in the nucleus before sent to the cytoplasm. b. Both ends of the primary RNA transcript are altered and certain interior sections of the RNA are cut out. c. The end result is a mature mRNA molecule ready for translation. 28. RNA PROCESSING: Ends of mRNA a. Each end is modified in a unique way. b. 5 end receives a cap made of a modified guanine nucleotide. c. An enzyme adds 50 to 250 adenines following the AAUAAA signal called the poly-A-tail. d. Both will help export mature mRNA from nucleus, help protect mRNA from degradation, help ribosomes attach to mRNA. E. UTR: 3 and 5 untranslated regions not translated into proteins but participate in binding to ribosome. 29. RNA SPLICING a. Cut-and-paste processing in the nucleus removing large portions of non-coding stretches of nucleotides, intervening sequences, introns. b. Human DNA transcription unit is ~27,000 bp. It only takes 1,200 nucleotides in RNA to code for the average size protein of 400 amino acids. c. Coding regions are called exons. 30. SPLICEOSOME a. A short nucleotide sequence at each end of the intron signal RNA splicing. b. Several different snRNPs (snurps) join proteins to form a spliceosome.
c. The spliceosome releases the intron and connects the flanking exons. 31. WHY INTRONS AND EXONS? Alternative DNA splicing! a. Thought to be the reason humans can get away with a relatively small number of genes, about 1 times as many as a fruit fly. b. Because of alternative splicing, the number of different protein products an organism produces can be much greater than its number of genes. 32. TRANSLATION CLOSE UP A. Ribosome: In and Out Burgers drive-thru (organelle). B. mRNA: Individuals (messengers) ordering burgers or polypeptides (proteins). C. tRNA: trucks that deliver the materials (amino acids) to fill the multiple burger (polypeptide) order. 33. tRNA ANTICODON a. Each tRNA is used repeatedly; ~80 nucleotides long, and is actually 3 dimensional. 34. Aminoacyl-tRNA synthetases join amino acids to a tRNA. a. Enzymes and ATP at work AGAIN! b. There are 20 different synthetases, one for each (redundant type) of the amino acids. c. It is an endergonic process to join the tRNA and amino acid together through the hydrolysis and energy donation of ATP. 35. WOBBLE a. There are ~45 tRNAs for the 61 possible amino acids to match up with the mRNA. b. Versatility possible because base-pairing rules are relaxed between the third base of a codon and the corresponding base of a tRNA anticodon. c. In very technical terms this is called wobble. 36. WOBBLE a. Wobble explains why the synonymous codons for a given amino acid can differ in their third base (look at the genetic code) but usually not in their other bases. 37. RIBOSOMES AND THE INITIATION OF TRANSLATION a. Ribosomes made up of two subunits named the large and small subunit, constructed of proteins and RNA molecules called ribosomal RNAs (rRNA). b. The small ribosomal subunit binds to the molecule of mRNA. c. The arrival of the large subunit completes the initiation complex. The start codon establishes the reading frame. d. mRNA binding site, 3 tRNA binding sites: E, P, A 38. ELONGATION IN TRANSLATION a. The mRNA is moved through the ribosome in one direction only, the 5 end first. The ribosome and the mRNA move relative to each other, uni-directionally, codon by codon. b. Codon recognition at the A site. c. Peptide bond formation at the P site. d. The ribosome translocates the tRNA in the A site to the P site, and simultaneously moves the empty tRNA to the E site to exit. 39. TERMINATION FINALLY! a. Elongation continues until a stop codon in the mRNA reaches the A site of the ribosome.
b. Protein called release factor binds to the A site. It causes the addition of a water molecule instead of an amino acid, which hydrolyzes the bond, releasing the polypeptide. Goodbye. 40. POLYRIBOSOMES a. An mRNA molecule is generally translated SIMULTANEOUSLY by several ribosomes in clusters called polyribosomes (or polysomes). b. Seen with an electron microscope. c. Found in prokaryotes and eukaryotes. 41. SUMMARY OF TRANSCRIPTION AND TRANSLATION
You might also like
- The Yan ReportDocument26 pagesThe Yan ReportZerohedge96% (117)
- How Genes Work GENE - Biological Unit of HeredityDocument5 pagesHow Genes Work GENE - Biological Unit of HeredityAdonis Besa100% (1)
- Genetic Control of Protein Structure & FunctionDocument60 pagesGenetic Control of Protein Structure & Functionathirah100% (1)
- C Science 10 Quarter 3 Module 3 (Week 4)Document14 pagesC Science 10 Quarter 3 Module 3 (Week 4)Daisy Soriano PrestozaNo ratings yet
- K101PracEx4 SP19 KeyDocument10 pagesK101PracEx4 SP19 KeyBraxton PhillipsNo ratings yet
- Study Guide Protein Synthesis (KEY)Document2 pagesStudy Guide Protein Synthesis (KEY)owls_1102No ratings yet
- DNA Blueprint of Life ExplainedDocument12 pagesDNA Blueprint of Life ExplainedhanzNo ratings yet
- 6 Genetics PDFDocument97 pages6 Genetics PDFasaNo ratings yet
- Protein SynthesisDocument135 pagesProtein SynthesisCarlaNo ratings yet
- Cell Final ExamDocument8 pagesCell Final ExamJorge GomezNo ratings yet
- Genetic Materia Medica Tri Miasmatic Materia Medica Prafull Vijayakar.07909 2introductionDocument5 pagesGenetic Materia Medica Tri Miasmatic Materia Medica Prafull Vijayakar.07909 2introductionRashmi MishraNo ratings yet
- HHMI - The p53 Gene and CancerDocument4 pagesHHMI - The p53 Gene and CancerThe vegetal saiyanNo ratings yet
- Final Exam StsDocument65 pagesFinal Exam StsChoe Yoek Soek0% (1)
- Chromosomal AbnormalitiesDocument3 pagesChromosomal AbnormalitiesYra Mie DitaroNo ratings yet
- The Importance of DNA and RNADocument13 pagesThe Importance of DNA and RNAJamieNo ratings yet
- Biology NIOSDocument927 pagesBiology NIOSApshar RizwanNo ratings yet
- Chapter Quiz General Biology 2Document7 pagesChapter Quiz General Biology 2Nikki San GabrielNo ratings yet
- MCQSDocument10 pagesMCQSahmed100% (2)
- Unit IV Lecture Notes 2019 RevisedDocument6 pagesUnit IV Lecture Notes 2019 RevisedSteve SullivanNo ratings yet
- Biological PhysicsDocument12 pagesBiological Physicsucing_330% (1)
- MCQ MolecularDocument11 pagesMCQ MolecularMahmOod Gh100% (2)
- Structural and Evolutionary Genomics: Natural Selection in Genome EvolutionFrom EverandStructural and Evolutionary Genomics: Natural Selection in Genome EvolutionRating: 5 out of 5 stars5/5 (1)
- Multiple Choice Questions in Molecular BiologyDocument72 pagesMultiple Choice Questions in Molecular Biologydanishuddin_danish50% (2)
- BBC - Higher Bitesize Biology - RNA and Protein Synthesis - PrintDocument8 pagesBBC - Higher Bitesize Biology - RNA and Protein Synthesis - PrintGrace AngelNo ratings yet
- CH 13 RNA and Protein SynthesisDocument12 pagesCH 13 RNA and Protein SynthesisHannah50% (2)
- Dhiraj Kumar, Chengliang Gong-Trends in Insect Molecular Biology and Biotechnology-Springer International Publishing (2018) PDFDocument376 pagesDhiraj Kumar, Chengliang Gong-Trends in Insect Molecular Biology and Biotechnology-Springer International Publishing (2018) PDFAndres Felipe Arias MosqueraNo ratings yet
- Biology Today and Tomorrow Without Physiology 5th Edition Starr Solutions Manual 1Document10 pagesBiology Today and Tomorrow Without Physiology 5th Edition Starr Solutions Manual 1sook100% (32)
- Biology Today and Tomorrow Without Physiology 5th Edition Starr Solutions Manual 1Document14 pagesBiology Today and Tomorrow Without Physiology 5th Edition Starr Solutions Manual 1janesilvapwoikcfjgb100% (25)
- CHAP 5 GeneticsDocument27 pagesCHAP 5 GeneticsAhmed AliNo ratings yet
- TRANSLATIONDocument28 pagesTRANSLATIONDaryl FCNo ratings yet
- Binder Key 2010-DnaDocument15 pagesBinder Key 2010-Dnaapi-292000448No ratings yet
- Bio Practice 7.2Document24 pagesBio Practice 7.2Xian Ni WooNo ratings yet
- CHAPTER 12 Protein SynthesisDocument10 pagesCHAPTER 12 Protein SynthesisNadeem IqbalNo ratings yet
- Protein Synthesis Transcription and Translation Distance LearningDocument10 pagesProtein Synthesis Transcription and Translation Distance LearningjaneyzhouNo ratings yet
- Biological Science Canadian 2Nd Edition Freeman Solutions Manual Full Chapter PDFDocument36 pagesBiological Science Canadian 2Nd Edition Freeman Solutions Manual Full Chapter PDFlewis.barnes1000100% (12)
- Chapter 5 - Expression of Biological InformationDocument3 pagesChapter 5 - Expression of Biological InformationNur HanisNo ratings yet
- Class Xii Biology Chapter 6 - Molecular Basis of InheritanceDocument8 pagesClass Xii Biology Chapter 6 - Molecular Basis of InheritanceAkshit Sharma VIII Diasy Roll no 6No ratings yet
- BIL 255 Exam Questions on DNA-Protein SynthesisDocument31 pagesBIL 255 Exam Questions on DNA-Protein SynthesisMax ArmourNo ratings yet
- De La Proteina La GenaDocument67 pagesDe La Proteina La GenaiuventasNo ratings yet
- Essentials of The Living World 5th Edition George Johnson Solutions ManualDocument5 pagesEssentials of The Living World 5th Edition George Johnson Solutions Manualjuliemcintyretzydiengcr100% (26)
- DNA vs RNA: A Comparison of Structures and FunctionsDocument4 pagesDNA vs RNA: A Comparison of Structures and Functionsmarianna hutchisonNo ratings yet
- BMM GM2 ADocument6 pagesBMM GM2 ASleepyHead ˋωˊNo ratings yet
- Exam 3 RetakeDocument8 pagesExam 3 RetakeMahima SikdarNo ratings yet
- Full Download Biotechnology 2nd Edition Clark Test BankDocument32 pagesFull Download Biotechnology 2nd Edition Clark Test Bankhildasavardpro100% (32)
- DNA and Protein SynthesisDocument34 pagesDNA and Protein SynthesisJessa Mae Infante PadillaNo ratings yet
- Microbiology A Human Perspective 7th Edition Nester Test BankDocument40 pagesMicrobiology A Human Perspective 7th Edition Nester Test Bankmagnusngah7aaz100% (28)
- Unit IV Lecture NotesDocument5 pagesUnit IV Lecture NotesSteve SullivanNo ratings yet
- Human Anatomy & Physiology I - Dr. Sullivan Unit IV - Cellular Function Chapter 4, Chapter 27 (Meiosis Only)Document5 pagesHuman Anatomy & Physiology I - Dr. Sullivan Unit IV - Cellular Function Chapter 4, Chapter 27 (Meiosis Only)Steve SullivanNo ratings yet
- The Central Dogma of Molecular BiologyDocument28 pagesThe Central Dogma of Molecular BiologysannsannNo ratings yet
- Pyq Cbse 2021-1-2Document19 pagesPyq Cbse 2021-1-2Yuvraj MehtaNo ratings yet
- Chapter 13 Test A RNA and Protein Synthesis ANSWERS PDFDocument6 pagesChapter 13 Test A RNA and Protein Synthesis ANSWERS PDFxspiiirONo ratings yet
- Long Test (Protein Synthesis)Document2 pagesLong Test (Protein Synthesis)ArdhieeNo ratings yet
- Finals Examination Biochemistry Lecture RNA Translation and MutationDocument7 pagesFinals Examination Biochemistry Lecture RNA Translation and MutationAlbertNo ratings yet
- Full Download Genetics and Genomics in Nursing and Health Care 1st Edition Beery Test BankDocument35 pagesFull Download Genetics and Genomics in Nursing and Health Care 1st Edition Beery Test Bankbeliechopper05srz100% (38)
- Protein SynthesisDocument35 pagesProtein SynthesisDaryl GumacalNo ratings yet
- Dwnload Full Genetics and Genomics in Nursing and Health Care 1st Edition Beery Test Bank PDFDocument35 pagesDwnload Full Genetics and Genomics in Nursing and Health Care 1st Edition Beery Test Bank PDFzaridyaneb100% (10)
- Genetics Chapter 8Document19 pagesGenetics Chapter 8Zahraa TermosNo ratings yet
- Dna, Rna and Protein SynthesisDocument26 pagesDna, Rna and Protein SynthesisB R YNo ratings yet
- Rna PDFDocument49 pagesRna PDFbangbro93_30900715No ratings yet
- DNA, RNA, Protein Synthesis ReviewDocument3 pagesDNA, RNA, Protein Synthesis ReviewChassy KammyNo ratings yet
- Bacterial Genetics in 40 CharactersDocument14 pagesBacterial Genetics in 40 Charactersعلي الكوافيNo ratings yet
- Central Dogma FlowDocument8 pagesCentral Dogma FlowAvirup RayNo ratings yet
- What Is The Main Function of DNA TranscriptionDocument8 pagesWhat Is The Main Function of DNA TranscriptionMega XtericsNo ratings yet
- Amino Acid Polypeptide End Product TranslationDocument5 pagesAmino Acid Polypeptide End Product TranslationAcep AbdullahNo ratings yet
- Dogma Sentral Dalam Biologi Kuliah23 OktoberDocument25 pagesDogma Sentral Dalam Biologi Kuliah23 OktoberararapiaNo ratings yet
- Heredity Protein SynthesisDocument23 pagesHeredity Protein SynthesispaulangelomicoNo ratings yet
- Genetic Determinisim JournalDocument11 pagesGenetic Determinisim JournalDave RkdNo ratings yet
- Bekish-VY - Multiple Choice Questions Book On Medical Biology - 2008Document78 pagesBekish-VY - Multiple Choice Questions Book On Medical Biology - 2008nishaninishaNo ratings yet
- Assign 01 (8610) Wajahat Ali Ghulam BU607455 B.ed 1.5 YearsDocument9 pagesAssign 01 (8610) Wajahat Ali Ghulam BU607455 B.ed 1.5 YearsAima Kha KhanNo ratings yet
- Genetics exam matching questionsDocument15 pagesGenetics exam matching questionsElizabeth LeonNo ratings yet
- DNA Structure Reveals Genetic CodeDocument77 pagesDNA Structure Reveals Genetic CodeCoco LocoNo ratings yet
- Genetic and ChromosomesDocument16 pagesGenetic and ChromosomesJay PornelaNo ratings yet
- Science Syllabi 07-08Document177 pagesScience Syllabi 07-08Varun KurtkotiNo ratings yet
- GenBio2 Q3 M4 6 Learner PDFDocument48 pagesGenBio2 Q3 M4 6 Learner PDFFlores Carl JosephNo ratings yet
- Saint Catherine Academy Earth and Life Science Final ExamDocument3 pagesSaint Catherine Academy Earth and Life Science Final ExamJanine Ginog FerrerNo ratings yet
- AP Biology Lab 6 TransformationDocument3 pagesAP Biology Lab 6 TransformationLogan GreenNo ratings yet
- Reproduction and Meiosis MarkschemeDocument16 pagesReproduction and Meiosis Markschememahek hNo ratings yet
- GREDocument7 pagesGREBoris ShalomovNo ratings yet
- Genetics 22-23Document9 pagesGenetics 22-23hiNo ratings yet
- NCERT Solutions For Class 12 Biology Chapter 6 Molecular Basis of InheritanceDocument10 pagesNCERT Solutions For Class 12 Biology Chapter 6 Molecular Basis of InheritanceSneha HipparkarNo ratings yet
- Reporter Gene Assay Slides 2021Document14 pagesReporter Gene Assay Slides 2021kittyngameNo ratings yet
- Cap 1 Parasitic NematodeDocument24 pagesCap 1 Parasitic NematodeJorge Chavez FloresNo ratings yet
- Isolation and Identification of Cytrid From Mangrove AreaDocument4 pagesIsolation and Identification of Cytrid From Mangrove AreaFaiqNo ratings yet
- RNAProtein Synthesis SEDocument7 pagesRNAProtein Synthesis SEAmbrielle WhiteNo ratings yet
- Anthro Unit 10Document30 pagesAnthro Unit 10Sanjana IASNo ratings yet
- Epilepsia EpicongressDocument245 pagesEpilepsia EpicongressВасилий КоптеловNo ratings yet