Professional Documents
Culture Documents
How To Sequence A Protein: W. Robert Midden Department of Chemistry Bowling Green State University
Uploaded by
scropion_78Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
How To Sequence A Protein: W. Robert Midden Department of Chemistry Bowling Green State University
Uploaded by
scropion_78Copyright:
Available Formats
How To Sequence
A Protein
W. Robert Midden
Department of Chemistry
Bowling Green State University
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Protein Sequencing
Preliminary Steps
For multisubunit proteins, the individual
protein chains must first be separated
Break interchain disulfide bonds, if necessary
Two reagents are commonly used:
performic acid
mercaptoethanol
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
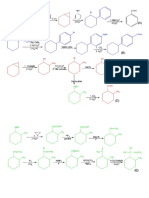
2-Mercaptoethanol
OO OO OO OO
HH HH
CC NN CC CC CC NN CC CC
HH HH
CH
CH22 CH
CH22
SS HHSS CH
CH22 CH
CH22 OH
OH SH
SH SS CH
CH22 CH
CH22 OH
OH
SS HS CH SH SS CH
CH22 CH
CH22 OH
HS CH22 CH
CH22 OH
OH SH OH
OO CH
CH22 OO OO CH
CH22 OO
CC NN CC CC CC NN CC CC
HH HH HH HH
2-mercaptoethanol reduces disulfides to sulfhydryls
But the sulfhydryls are easily oxidized back to the disulfide
3
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Preventing Reversal
OO OO OO OO
HH HH
CC NN CC CC CC NN CC CC
HH HH
CH OO CH
CH2 2 OO
CH
2
2
SH II CH2 CC
SS CH
CH2 2 CC
SH CH2
OO OO
OO OO OO OO
HH HH
CC NN CC CC CC NN CC CC
HH HH
CH CH
CH
CH
2
2
2
2
SH HH
2CC CH SS HH
2CC CH
CH2 2 CC NN
SH 2 CH CC NN
2
to prevent oxidation the suflhydryls are alkylated with
iodoacetic acid or acrylonitrile
4
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Perfomic Acid
OO OO OO OO
HH HH
CC NN CC CC CC NN CC CC
HH HH
CH CH
CH22
CH22
OO
SS SO
SO3-3-
HH CC OO OO HH
SS SO
SO3-3-
OO CH OO CH
CH22 OO
CH22 OO
CC NN CC CC CC NN CC CC
HH HH HH HH
Performic acid oxidizes cysteine to negatively charged
cysteic acid
5
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Reversal Prevented
OO OO
HH
CC NN CC CC
HH
CH
CH22 SO
SO3-3-
SO
SO3-3- OO CH
CH22 OO
CC NN CC CC
HH HH
The repulsion of the negatively charged SO3- groups
prevents reformation of the disulfide bond
Therefore alkylation is not necessary with performic acid
6
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Protein Sequencing
Preliminary Steps
After breaking disulfide bonds, the chains are
separated by disrupting noncovalent interchain
interactions with pH extremes, 8 M urea, 6 M
guanidium hydrochloride, or high salt
Then the individual protein chains are
separated by electrophoresis or chromatography
on the basis of size or charge
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Determining Amino
Acid Sequence
Once each protein is purified the amino acid
sequence is determined by:
1) determining the amino acid composition
(how many of each amino acid are in the
protein)
2) identifying the amino and carboxyl terminal
amino acids
3) cleaving the protein into two or more sets of
peptides using specific enzymatic or chemical
reagents such as trypsin or cyanogen bromide
8
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Determining
Protein Sequence
4) determining the amino acid sequence of each
of the peptide fragments
5) determining the entire protein sequence from
the sequences of overlapping peptide
fragments
6) locating the position of disulfide bridges
between cysteines
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Determining Amino
Acid Composition
The amino acid composition is determined by:
Hydrolysis with 6N HCl for one to three days
Separating and quantifying individual amino
acids by ion exchange HPLC using an amino
acid analyzer
10
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Determining the
N-Terminal Amino Acid
The N-terminal amino acid is determined using
either chemical reagents or enzymes
Chemical reagents include:
Sanger’s reagent
dansyl chloride
Edman Degradation
11
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Determining the
N-Terminal Amino Acid
Sanger’s reagent
Treat with dinitrofluorobenzene to
OO
+
OO form a dinitrophenyl (DNP)
N+N derivative of the amino-terminal
amino acid
Acid hydrolysis
Extract the DNP-derivative from the
OO
+
N+
acid hydrolysate with organic
N solvent
FF OO Identify the DNP-derivative by
chromatography and comparison
with standards
12
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Determining the
N-Terminal Amino Acid
HH3CC CH
Dansyl chloride
CH33
3
NN (dimethylaminonaphthylenesulfonyl
chloride)
Forms a highly fluorescent derivative
of the amino-terminal amino acid
Identified by chromatography and
fluorescence detection after acid
OO SS OO
hydrolysis
Highly senstive
ClCl
Best choice when the amount of
protein is limited
13
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Determining the
N-Terminal Amino Acid
Edman degradation
phenylisothiocyanate (phenyl-N=C=S) adds to N-terminus
then acid treatment cleaves the N-terminal amino acid as a
PTH derivative
the remaining protein chain is intact and the cycle can be
repeated
under ideal conditions the sequence of 30-60 amino acids
can be determined
Leucine aminopeptidase
enzyme from hog kidney hydrolyzes the N-terminal
peptide bond
best with nonpolar amino acids
14
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Determining the
C-Terminal Amino Acid
Hydrazinolysis
hydrazine at 100°C cleaves all peptide bonds forming
hydrazides except for the carboxyl terminal
C-terminus reduced with LiAlH4
forms amino alcohol at C-terminus
Carboxypeptidases
enzymatic removal of C-terminus
Carboxypeptidase A all except proline, arginine and lysine
Carboxypeptidase B only arginine and lysine
Carboxypeptidase C any amino acid
care required since rate of removal varies with the type of
amino acid
15
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Peptide Fragments
After determining the amino acid composition
and the N & C-terminal amino acids, at least
two different sets of protein fragments are
needed for sequencing
16
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Why Use Fragments?
Why is the protein broken into fragments? Why
isn’t the protein sequenced directly?
The sequencing methods currently available are
only accurate for peptides up to about 20-30
amino acids, 60 under ideal conditions
17
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Why 2 Sets of
Fragments?
Why can't the entire protein amino acid
sequence be determined from a single set of
peptide fragments obtained by cleavage with a
single reagent?
There’s no way to determine how the fragments
are connected with just one set
A second or third set of fragments are used to
deduce how the fragments are connected by
identification and comparison of overlapping
sequnces
18
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Protein Cleavage
Reagents
What types of reagents are best suited for
preparing these sets of fragments?
Reagents that cleave the protein chain only at a
few specific sites forming fragments that are
less than 20-30 amino acids in length
19
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Protein Cleavage
Reagents
Chemical or enzymatic reagents can be used to
prepare protein fragments
The most commonly used reagents are:
cyanogen bromide
various enzymes including
trypsin
chymotrypsin
clostripain
Staphylococcal protease
various endopeptidases
20
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Cyanogen Bromide
CH
CH33 Br
Br CH
CH33 HH3CC
3
+
SS CC SS+ CC NN SS CC NN
CH NN CH
CH22 CH
CH22 CH22
CH CH
CH22 OO
CH22 OO HH2CC
2
OO
NN CC CC NN NN CC CC NN NN CC CC
OO
HH HH HH HH HH HH HH HH
At which amino acid in the protein sequence does the
reagent, cyanogen bromide, cleave protein chains?
At internal methionines by reaction with the methionine
sulfur as illustrated above
21
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Trypsin & Chymotrypsin
Where in the protein sequence do the enzymes,
trypsin and chymotrypsin cleave protein
chains?
trypsin cleaves at the carboxyl side of amino
acids with positively charged side chains such
as lysine and arginine
chymotrypsin cleaves at the carboxyl side of
amino acids with aromatic side chains such as
phenylalanine and tyrosine
22
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Clostripain
Where in the protein sequence does the
enzyme, clostripain, cleave?
prefers positively charged amino acids, arginine
even more than lysine
narrower specificity than tryptophan
which enzyme is likely to produce larger
fragments?
23
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Staphyloccal Protease
Where in the protein sequence does the
enzyme, Staphylococcal protease cleave?
carboxyl side of acidic amino acids in
phosphate buffer
in acetate or bicarbonate buffer it is more
specific and cleaves only glutamic acid
24
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Endopeptidases
The following endopeptidases are less specific
than the enzymes metioned above
Pepsin, papain, subtilisin, thermolysin, elastase
(papain is the active ingredient in meat tenderizer, soft
contact cleansing solutions, some laundry detergents)
These enzymes are most often used to further
reduce the size of large tryptic or chymotryptic
fragments
25
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
How are Peptide
Fragments Separated?
Usually by column chromatography, often
HPLC
Separations are most often based on differences
in polarity (reverse phase) or electric charge
(ion exchange)
26
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Edman Degradation
Edman degradation is most often used to
sequence the peptides
It removes one amino acid from the N-terminal
end of the peptide during each cycle of the
procedure
The removal of the N-terminal amino acid is
accomplished using the reagent,
phenylisothiocyanate
27
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Edman Degradation
Pheylisothiocyanate attaches to the N-terminal
amino acid
The peptide amino nitrogen atom bonds to the
PITC carbon
Sulfur then bonds to the peptide carboxyl carbon
breaking the peptide bond
This cyclization forms a pheylthiohydantoin
derivative which is removed from the peptide
chain by treatment with anhydrous acid
Identified by extraction, treatment with aqueous
acid and analysis by chromatography
28
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Edman Degradation
NN
NH
NH
CC NN
OO SS
CC SS
HH
SS
HHNN NN
HH2NN
2 HH
3CC HH
HH CC CH
CH3 3 3
HH CC CH
CH
3
3
CC OO
CC OO HH
2NN
2
NH
NH
NH
NH
29
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Disulfide Bridges
The location of disulfide bridges can be
determined by diagonal electrophoresis
Fragments with intact disulfide bonds are
electrophoresed in one dimension
Treated with fumes of performic acid to cleave
disulfide bonds
Then electrophoresed in the second dimension
Fragments that had no disulfide bonds will be on
the diagonal
Fragments that had disulfide bonds will migrate
off diagonal due to altered mobility
30
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Mass Spectroscopy
Used for sequencing peptides
Peptides are fragmented in the mass
spectrometer
The fragments are identified by their
mass/charge ratio
Peptide mixtures can be analyzed using a
temperature gradient
The temperature gradient causes variation in
signals corresponding to different peptides
31
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Protein Sequencing
by DNA Sequencing
In fact, while you have just learned how to
sequece a protein by chemical and enzymatic
degradation, protein sequences are now most
often determined by translating the
corresponding cloned genes
This latter process is usually easier and quicker
once the gene corresponding to a given protein
has been identified
32
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
Sequence Databases
International databases of protein sequences
are maintained
Many of these databases are accessible via the
internet
Examples:
GenBank
Protein Identification Resource (PIR)
European Molecular Biology Data Library
(EMBL)
33
Copyright © 1998 W.R. Midden
All Rights Reserved Bowling Green State University Protein Sequencing
You might also like
- Phthalic2 B PDFDocument11 pagesPhthalic2 B PDFIzzati AhmadNo ratings yet
- Selection Strategy of Mechanical SealDocument7 pagesSelection Strategy of Mechanical Sealdhanu_aquaNo ratings yet
- Naming Alkenes WorksheetDocument2 pagesNaming Alkenes WorksheetJaya Chitra Degala Ramalu100% (1)
- Naming Alkanes - Alkenes and AlkynesDocument6 pagesNaming Alkanes - Alkenes and AlkynesCarla SanaNo ratings yet
- IB QBANK 6.1 - p1-p2Document35 pagesIB QBANK 6.1 - p1-p2Naz Gümüşlüoğlu100% (1)
- Synthesis of Acetyl Salicylic AcidDocument5 pagesSynthesis of Acetyl Salicylic AcidSilvia AryaniNo ratings yet
- 3 Naming Alkynes Ws KeyDocument2 pages3 Naming Alkynes Ws KeyJaya Chitra Degala RamaluNo ratings yet
- H NMR Problems: - How Many Unique Proton Environments Are There inDocument26 pagesH NMR Problems: - How Many Unique Proton Environments Are There inFatima AhmedNo ratings yet
- How To Sequence A Protein: W. Robert Midden Department of Chemistry Bowling Green State UniversityDocument33 pagesHow To Sequence A Protein: W. Robert Midden Department of Chemistry Bowling Green State UniversitySophia GoNo ratings yet
- 22BSD7019Document2 pages22BSD7019SAI CHARAN KOTAPATINo ratings yet
- Molecular RearrangementsDocument29 pagesMolecular RearrangementsThabiso GwijiNo ratings yet
- 5lkmq-Acides AminesDocument1 page5lkmq-Acides Aminessoumailakaila59No ratings yet
- Chela Tesch Elating Agents 2017Document3 pagesChela Tesch Elating Agents 2017Sharin Bin Ab GhaniNo ratings yet
- NH CH CO H: 6.11 Amino Acids, Proteins and DNADocument14 pagesNH CH CO H: 6.11 Amino Acids, Proteins and DNAPedro Moreno de SouzaNo ratings yet
- Kukdo Epoxy Resins & Hardeners Classification GuideDocument38 pagesKukdo Epoxy Resins & Hardeners Classification GuideAkhtar aliNo ratings yet
- Organic compounds identificationDocument5 pagesOrganic compounds identificationPFENo ratings yet
- 0331 S 05 BioenergyDocument44 pages0331 S 05 BioenergyDaisyNo ratings yet
- AldeDocument2 pagesAlde14.Hajjan MNo ratings yet
- Sample Name: Thiol Terminated Polystyrene Sample # P18811-SSHDocument1 pageSample Name: Thiol Terminated Polystyrene Sample # P18811-SSHOscar PiñeresNo ratings yet
- Biological Molecules - ProteinsDocument18 pagesBiological Molecules - ProteinsblackmoneygrabberNo ratings yet
- Quimica 11 - Didáctica MultimediaDocument197 pagesQuimica 11 - Didáctica MultimediaviviarcelopezNo ratings yet
- Photo ChemistryDocument26 pagesPhoto ChemistryAkowuah SamuelNo ratings yet
- For Each of The Following Reactions, Give The Structure of The Product and Indicate Whether The Mechanism Is Likely To Be SN1, SN2, Both or NeitherDocument2 pagesFor Each of The Following Reactions, Give The Structure of The Product and Indicate Whether The Mechanism Is Likely To Be SN1, SN2, Both or NeitherVarokah VarNo ratings yet
- HW - CHAPTER 09 - Trương Chí Đ I - 2052935Document5 pagesHW - CHAPTER 09 - Trương Chí Đ I - 2052935Hidai Truc NgoNo ratings yet
- Basics of Photochemistry and Norrish Type I ReactionDocument12 pagesBasics of Photochemistry and Norrish Type I Reactionnidhi vashisthaNo ratings yet
- Alcohol Nomenclature - Summative AssessmentDocument2 pagesAlcohol Nomenclature - Summative AssessmentDiana Carolina DuarteNo ratings yet
- Alcohol Nomenclature - Summative AssessmentDocument2 pagesAlcohol Nomenclature - Summative AssessmentDiana Carolina DuarteNo ratings yet
- (S.C. Rastogi) Essentials of Animal Physiology, 4t (BookSee - Org) 246Document1 page(S.C. Rastogi) Essentials of Animal Physiology, 4t (BookSee - Org) 246Indah Rizka AprilianiNo ratings yet
- DM pp21-40Document20 pagesDM pp21-40MLUNGISI MkhwanaziNo ratings yet
- Answers To AssignmentDocument1 pageAnswers To AssignmentIgbereyivwe TejiriNo ratings yet
- Amino Acids Metabolism &: Urea CycleDocument11 pagesAmino Acids Metabolism &: Urea Cyclemuhammad amjadNo ratings yet
- Drug Metabolism: Prepared By: Alfonso Cantor, RPHDocument18 pagesDrug Metabolism: Prepared By: Alfonso Cantor, RPHJericSalcedoNo ratings yet
- 5.111 Principles of Chemical Science: Mit OpencoursewareDocument7 pages5.111 Principles of Chemical Science: Mit OpencoursewareAgung SujatmikoNo ratings yet
- i) 2,2-Dimethylbutane(ii) 3-Ethyl-2-methylhexane (iii) 2-Methyl-5-ethylnonaneDocument45 pagesi) 2,2-Dimethylbutane(ii) 3-Ethyl-2-methylhexane (iii) 2-Methyl-5-ethylnonaneAnubhab100% (2)
- Chemistry PracticeDocument13 pagesChemistry PracticeSiddharth KrishnamurthyNo ratings yet
- Synthetic Organic Chemistry 932 Jalal Zahra Homework 1Document1 pageSynthetic Organic Chemistry 932 Jalal Zahra Homework 1محمد مصطفىNo ratings yet
- Biological MembranesDocument15 pagesBiological MembranesTazinNo ratings yet
- Amino Acids NotesDocument17 pagesAmino Acids NotesNguyễn SunNo ratings yet
- Macam-Macam Asam AminoDocument2 pagesMacam-Macam Asam Aminoarif nur rokhmanNo ratings yet
- 138dre - pdf23 12 12 14 10 34 778Document10 pages138dre - pdf23 12 12 14 10 34 778bdaljbarrhmany4No ratings yet
- Topic 17 Exercise 1 - Naming Organic Compounds: C CL ODocument1 pageTopic 17 Exercise 1 - Naming Organic Compounds: C CL OAmmaarah PatelNo ratings yet
- đọc tênDocument7 pagesđọc tênthái đức thắngNo ratings yet
- Lipida: Indah Saraswati, M. SCDocument55 pagesLipida: Indah Saraswati, M. SCPutriNo ratings yet
- Presentación 1Document2 pagesPresentación 1liz8aknoNo ratings yet
- PH-6 - Mains - Answers - ChemistryDocument17 pagesPH-6 - Mains - Answers - Chemistrytanu15048No ratings yet
- Using IR To Solve ProblemsDocument21 pagesUsing IR To Solve ProblemsIka SanjiwaniNo ratings yet
- AnestheticsDocument67 pagesAnestheticsEsha pantNo ratings yet
- Assignment 1Document12 pagesAssignment 1Tshiamo MotaungNo ratings yet
- Hydrocarbon Nomenclature WorksheetDocument3 pagesHydrocarbon Nomenclature WorksheetCaseelyn Joy NantizaNo ratings yet
- GLUCIDELEDocument25 pagesGLUCIDELEDisneyNo ratings yet
- Naglaa Ibrahim Mohamed Azab - Protein MetabolismDocument80 pagesNaglaa Ibrahim Mohamed Azab - Protein MetabolismIbi Yulia SetyaniNo ratings yet
- CH12 Unit04 AOSRA02Document7 pagesCH12 Unit04 AOSRA02PyNo ratings yet
- 13 Organic ChemistryDocument13 pages13 Organic ChemistryTo SecretNo ratings yet
- Alkenes and Alkynes Worksheet PDFDocument3 pagesAlkenes and Alkynes Worksheet PDFDoha Hosam Shalaby HamamNo ratings yet
- Amino Acid Residue Mass and Proton AffinityDocument2 pagesAmino Acid Residue Mass and Proton AffinityMichelle FactoNo ratings yet
- Adobe Scan 30-Aug-2022Document5 pagesAdobe Scan 30-Aug-2022Harshit RajNo ratings yet
- L9 Neighbouring Group ParticipationDocument32 pagesL9 Neighbouring Group Participationvanwani.mozeelNo ratings yet
- Alkene Reaction Practice MechanismsDocument6 pagesAlkene Reaction Practice MechanismsJoe JulianNo ratings yet
- ALCOHOL : AlcoholsDocument11 pagesALCOHOL : AlcoholsSamirNo ratings yet
- Chemical structures and IUPAC names of organic compoundsDocument2 pagesChemical structures and IUPAC names of organic compoundsCamil SolerNo ratings yet
- AntihypertensiveDocument45 pagesAntihypertensiveapt pkmarcamanikNo ratings yet
- Cadeias CarbonicasDocument1 pageCadeias CarbonicasGislaine Cavassim GemelliNo ratings yet
- Danyar Et Al., 2020 - FinalDocument24 pagesDanyar Et Al., 2020 - FinalSardar SaleemNo ratings yet
- Clariant - Exolit Overview 2016Document12 pagesClariant - Exolit Overview 2016xy2zjgNo ratings yet
- Module 1 - Clinical BacteDocument3 pagesModule 1 - Clinical BacteElyssa VergaraNo ratings yet
- B.sc. ChemistryDocument66 pagesB.sc. ChemistryDr. Dinesh KumarNo ratings yet
- Biol 1362 Lab 2Document3 pagesBiol 1362 Lab 2kittypowerNo ratings yet
- JEE Test Series Answers for Physics, Chemistry and MathsDocument10 pagesJEE Test Series Answers for Physics, Chemistry and MathsANUNo ratings yet
- Physicochemical Properties Estimation of Total Sugars and Vitamin C Content of Pomegranate Cultivars Arakta and GanseshDocument6 pagesPhysicochemical Properties Estimation of Total Sugars and Vitamin C Content of Pomegranate Cultivars Arakta and GanseshNemes BiancaNo ratings yet
- CHY2023 - Unit 3 Aromatic HydrocarbonsDocument75 pagesCHY2023 - Unit 3 Aromatic HydrocarbonsZhori Duberry100% (1)
- Chemical Tests PDFDocument2 pagesChemical Tests PDFSyafiqah ArinaNo ratings yet
- Enzymes: Handbook: MetabolismDocument2 pagesEnzymes: Handbook: Metabolismsmol ukeleleNo ratings yet
- CableDocument24 pagesCableGnanesh GNo ratings yet
- Furmanite T233 300115Document3 pagesFurmanite T233 300115mujahidyousafNo ratings yet
- Jamb 2024 MockDocument20 pagesJamb 2024 Mockebubeonubogu2005No ratings yet
- Nucleic Acids QuestionsDocument4 pagesNucleic Acids QuestionsShubham GhoshNo ratings yet
- Postharvest Physiology 4th YearDocument54 pagesPostharvest Physiology 4th YearYobsan BushuraNo ratings yet
- Arrhenius Bronsted Lewis Acids: Acids: AcidsDocument3 pagesArrhenius Bronsted Lewis Acids: Acids: AcidsBianca Del RosarioNo ratings yet
- Development of Food Chemistry Natural Products and Nutrition ResearchDocument198 pagesDevelopment of Food Chemistry Natural Products and Nutrition ResearchRoberta MorganaNo ratings yet
- Organic Synthesis of VanillinDocument6 pagesOrganic Synthesis of VanillinSarah Alexander0% (1)
- Additive and Colour Preparations: For Extruded Polystyrene FoamsDocument22 pagesAdditive and Colour Preparations: For Extruded Polystyrene FoamsKarim HakimNo ratings yet
- Supercritical: Deposition MechanismsDocument6 pagesSupercritical: Deposition MechanismsTu LENo ratings yet
- Pullulanase Production and Usage in Food IndustryDocument8 pagesPullulanase Production and Usage in Food IndustryAbeer FatimaNo ratings yet
- Qualitative and Quantitative Tests For LipidsDocument33 pagesQualitative and Quantitative Tests For LipidsSharon GabrielNo ratings yet
- Practical Report: FHSC1214 Fundamentals of Cell Biology Foundation in ScienceDocument3 pagesPractical Report: FHSC1214 Fundamentals of Cell Biology Foundation in ScienceEngNo ratings yet
- Fundamentals of Biochemistry Life at The Molecular Level 5Th Edition Voet Test Bank Full Chapter PDFDocument50 pagesFundamentals of Biochemistry Life at The Molecular Level 5Th Edition Voet Test Bank Full Chapter PDFgillanolympia4091253100% (10)
- Trends in Food Science & Technology: Mahmood Alizadeh-Sani, Esmail Mohammadian, Jong-Whan Rhim, Seid Mahdi JafariDocument52 pagesTrends in Food Science & Technology: Mahmood Alizadeh-Sani, Esmail Mohammadian, Jong-Whan Rhim, Seid Mahdi JafaritriNo ratings yet
- Group 7 Toxicology - Ozone Layet DepletionDocument18 pagesGroup 7 Toxicology - Ozone Layet DepletionNurul IsmailNo ratings yet