Professional Documents
Culture Documents
Prokaryotes Vs Eukaryotes
Prokaryotes Vs Eukaryotes
Uploaded by
metzlogan84700 ratings0% found this document useful (0 votes)
22 views2 pagesProkaryotes and eukaryotes have differences in DNA replication, transcription, and translation. [1] DNA replication in prokaryotes initiates at a single origin with regulated initiation, while eukaryotes have thousands of origins and initiation factors. [2] Transcription in prokaryotes uses one RNA polymerase with a sigma factor for gene expression control, while eukaryotes have RNA polymerases I, II, and III. [3] Translation in prokaryotes uses 70S ribosomes and Shine-Delgarno sequences, while eukaryotes use 80S ribosomes, methyl-G capping, poly-A tails, and multiple initiation factors.
Original Description:
Test
Original Title
Prokaryotes vs Eukaryotes
Copyright
© © All Rights Reserved
Available Formats
DOCX, PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentProkaryotes and eukaryotes have differences in DNA replication, transcription, and translation. [1] DNA replication in prokaryotes initiates at a single origin with regulated initiation, while eukaryotes have thousands of origins and initiation factors. [2] Transcription in prokaryotes uses one RNA polymerase with a sigma factor for gene expression control, while eukaryotes have RNA polymerases I, II, and III. [3] Translation in prokaryotes uses 70S ribosomes and Shine-Delgarno sequences, while eukaryotes use 80S ribosomes, methyl-G capping, poly-A tails, and multiple initiation factors.
Copyright:
© All Rights Reserved
Available Formats
Download as DOCX, PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
22 views2 pagesProkaryotes Vs Eukaryotes
Prokaryotes Vs Eukaryotes
Uploaded by
metzlogan8470Prokaryotes and eukaryotes have differences in DNA replication, transcription, and translation. [1] DNA replication in prokaryotes initiates at a single origin with regulated initiation, while eukaryotes have thousands of origins and initiation factors. [2] Transcription in prokaryotes uses one RNA polymerase with a sigma factor for gene expression control, while eukaryotes have RNA polymerases I, II, and III. [3] Translation in prokaryotes uses 70S ribosomes and Shine-Delgarno sequences, while eukaryotes use 80S ribosomes, methyl-G capping, poly-A tails, and multiple initiation factors.
Copyright:
© All Rights Reserved
Available Formats
Download as DOCX, PDF, TXT or read online from Scribd
You are on page 1of 2
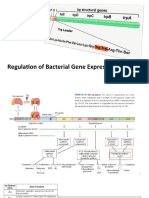
Prokaryotes Eukaryotes
DNA OriC Thousands of origins (ARS)
replication Initiation (regulated) ARS = ORC + ABF
Helicase Helicase
SSB RPA (~SSB)
Topoisomerase I Topoisomerase I/II
Gyrase (topo II) RNase H (~DNA pol I)
Primase DNA pol (initiate, primase)
DNA pol I (fill gap & remove DNA pol (repair)
primer) DNA pol (Mt.)
DNA pol II (repair) DNA pol (~pol III, lag)
DNA pol III (most of syn) DNA pol (~pol III, lead)
Ligase Ligase
Termination sequences Kinases start DNA rep Rep S-
phase
Pol complex (/+PCNA+RFC)
Telomerase
TSC No primer Initiation factors
First level of gene exp. Control RNA pol I (nucleolus, 45S
RNA pol (only 1) rRNA)
Core enzyme + factor RNA pol II (mRNA & miRNA)
No proofreading RNA pol III (5S rRNA & tRNA)
Promoter site (Pribnow) Promoter site
-35 TTGACA TATA box @ -30 (~-10)
-10 TATAAT Core factors: TF2A, TF2B
Topoisomerase (I & II) TF2D = TBP & TAFs
Rho dep/ind termination TF2H (helicase & kinase)
Excision repair
Polyadenylation
CPSF & CstF
TSL 50S + 30S = 70S 60S + 40S = 80S
Polycistronic mRNAs Monocistronic mRNAs
Shine Delgarno Seq. (purine Methyl-Gcap & poly A tail
rich) 9 total eIFs
IF1, IF2, IF3 (2,2B,3,4A,4B,4E,4G,5,6)
fMet-tRNAfMet Met-tRNAiMet
EF-Tu, EF-Ts, EF-G eEF-1a, eEF-1b, eEF-2
RF1 (UAA, UAG), RF2 (UAA, UGA), eRF-1, eRF-3
RF3
RNA-dependent DNA pol reverse transcriptase
Retroviruses, no proof reading, error prone
Mt DNA rep bootlace model
Includes complementary tRNA & mRNA to hybridize to the template
lagging strand
Mt TSC
Codes 13 proteins, 3 promoters, polycistronic transcripts tRNA rRNA
mRNA
You might also like
- MPC Bible DemoDocument19 pagesMPC Bible DemoFERA CENTRALNo ratings yet
- Postoperative Fever - UpToDate PDFDocument25 pagesPostoperative Fever - UpToDate PDFkatsuiaNo ratings yet
- 2012 Topics On The Dynamics of Civil StructuresDocument435 pages2012 Topics On The Dynamics of Civil Structuresaion_rebNo ratings yet
- Benjamin, Walter - Theses On The Philosophy of HistoryDocument11 pagesBenjamin, Walter - Theses On The Philosophy of HistoryAleksandra VeljkovicNo ratings yet
- Power Plant BasicsDocument80 pagesPower Plant Basicssandipchavan1280% (25)
- D CONSTITUTIONAL LAW The Philippines As A StateDocument13 pagesD CONSTITUTIONAL LAW The Philippines As A StateSunjeong Macho100% (1)
- Transcription and TranslationDocument35 pagesTranscription and TranslationMing mingNo ratings yet
- Central Dogma: Faculty of Medicine, Sriwijaya UniversityDocument51 pagesCentral Dogma: Faculty of Medicine, Sriwijaya UniversityIlsya PertiwiNo ratings yet
- Compre MolbioDocument93 pagesCompre MolbioDeniebev'z OrillosNo ratings yet
- Sash PlanesDocument5 pagesSash PlanesBlakdawg15100% (1)
- AVES de La Reserva PacayaDocument61 pagesAVES de La Reserva PacayaGianina J. Mundo PadillaNo ratings yet
- Chapter 4 Demolition WorksDocument25 pagesChapter 4 Demolition WorksAmir AlistyNo ratings yet
- TranscriptionDocument100 pagesTranscriptionSreshttNo ratings yet
- Introduction To Bioorganic Chemistry and Chemical Biology Ch9 AnswersDocument10 pagesIntroduction To Bioorganic Chemistry and Chemical Biology Ch9 AnswersJeremy Carter0% (1)
- 11 Chapter 5Document38 pages11 Chapter 5toobashafiNo ratings yet
- Pribnow 1975 Nucletide Sequence of RNA Polymerase Binding Site in T7 PromoterDocument5 pagesPribnow 1975 Nucletide Sequence of RNA Polymerase Binding Site in T7 PromoterAnaNo ratings yet
- Prokaryotes vs. EukaryotesDocument3 pagesProkaryotes vs. EukaryotesRichard HampsonNo ratings yet
- Transcription, Translation, Genetic Code Mutations (Part 2) - Compressed PDFDocument131 pagesTranscription, Translation, Genetic Code Mutations (Part 2) - Compressed PDFMuhammad Haris AshfaqNo ratings yet
- Transcription in EukaryotesDocument15 pagesTranscription in EukaryotesFenet AdamuNo ratings yet
- Biological Sequence Determination: ProteinDocument68 pagesBiological Sequence Determination: Proteinpathcell bioNo ratings yet
- RNA Synthesis and SplicingDocument64 pagesRNA Synthesis and SplicingLaurine PigossoNo ratings yet
- Biochemistry: RNA Synthesis and ProcessingDocument54 pagesBiochemistry: RNA Synthesis and ProcessingAqsa YaminNo ratings yet
- MODULE 1B SGD3 RNA Transcription and Translation An The Genetic CodeDocument9 pagesMODULE 1B SGD3 RNA Transcription and Translation An The Genetic CodeKristine Jade OdtujanNo ratings yet
- Transcription and TranslationDocument2 pagesTranscription and TranslationRobert Martin PuertaNo ratings yet
- Transcription and TranslationDocument2 pagesTranscription and TranslationRobert Martin PuertaNo ratings yet
- TranslationDocument1 pageTranslationGianne AbcedeNo ratings yet
- Gene ExpressionDocument46 pagesGene Expressionmohammed mahmoudNo ratings yet
- Biochemistry Kaplan (AutoRecovered)Document108 pagesBiochemistry Kaplan (AutoRecovered)Khoa VõNo ratings yet
- BIO353 Lecture13 (Regulation Prokaryotes) SHF 2021Document82 pagesBIO353 Lecture13 (Regulation Prokaryotes) SHF 2021kittyngameNo ratings yet
- RNA Structure, Functions WebDocument25 pagesRNA Structure, Functions WebEmad ManniNo ratings yet
- Dogma SentraDocument46 pagesDogma SentraindahonlyNo ratings yet
- Bioc 302 TableDocument7 pagesBioc 302 Table심재은No ratings yet
- LECTURE 12 TranslationDocument17 pagesLECTURE 12 TranslationAditi SharmaNo ratings yet
- MB Chapter 5 TranscriptionDocument32 pagesMB Chapter 5 TranscriptionMustee TeferaNo ratings yet
- Botany Chapter 1 (Cell)Document16 pagesBotany Chapter 1 (Cell)Peslegend TahasinNo ratings yet
- Transcription in Prokaryotes: Dindin H. Mursyidin Laboratory of Molecular Biology Lambung Mangkurat UniversityDocument39 pagesTranscription in Prokaryotes: Dindin H. Mursyidin Laboratory of Molecular Biology Lambung Mangkurat UniversityNadia Nur FitriaNo ratings yet
- Lecture 3Document6 pagesLecture 3tiaria.wilson24No ratings yet
- TranscriptionDocument72 pagesTranscriptionMurthy MandalikaNo ratings yet
- Dna L12 NotesDocument6 pagesDna L12 NotesellieNo ratings yet
- Analysis of Apicoplast Genome Sequence From Eimeria Maxima.: Nor Fazilah Ab Rahman and Kiew-Lian WanDocument1 pageAnalysis of Apicoplast Genome Sequence From Eimeria Maxima.: Nor Fazilah Ab Rahman and Kiew-Lian WanFazilah RahmanNo ratings yet
- Transcription in Prokaryotes: Single RNA PolymeraseDocument29 pagesTranscription in Prokaryotes: Single RNA PolymeraseShubhamNo ratings yet
- Protein Synthesis LegendDocument1 pageProtein Synthesis Legendkirby kirNo ratings yet
- Science of Living System: Arindam MondalDocument48 pagesScience of Living System: Arindam MondalSohini RoyNo ratings yet
- Study Guide Chapter 8Document4 pagesStudy Guide Chapter 8Fati UmaruNo ratings yet
- Prokaryotic Gene ExpressionDocument28 pagesProkaryotic Gene ExpressionNasiha el KarimaNo ratings yet
- Sintesis Protein Pada Prokariot: Evi Umayah UlfaDocument44 pagesSintesis Protein Pada Prokariot: Evi Umayah UlfaGrace IndriNo ratings yet
- Usanee Anukool (PH.D.) Clinical Microbiology Faculty of Associated Medical Sciences Chiang Mai University 2 July 2009Document60 pagesUsanee Anukool (PH.D.) Clinical Microbiology Faculty of Associated Medical Sciences Chiang Mai University 2 July 2009Suruk UdomsomNo ratings yet
- Gene Structure and Identification: Genes and Genomes Orfs and More Consensus Sequences Gene FindingDocument16 pagesGene Structure and Identification: Genes and Genomes Orfs and More Consensus Sequences Gene FindingimaemNo ratings yet
- Comparison of Transcription, Translation and ReplicationDocument1 pageComparison of Transcription, Translation and ReplicationDrashty DesaiNo ratings yet
- Chapter 2&3 Expression of Biological Information I and IIDocument17 pagesChapter 2&3 Expression of Biological Information I and IIRIFAH FIDZDIANI AIN BINTI RIFKIY FS21110415No ratings yet
- Lecture - 1Document33 pagesLecture - 1Bahesty Monfared AkashNo ratings yet
- Biomol R2 Kurniahtunnisa 0402517015 RefleksiDocument15 pagesBiomol R2 Kurniahtunnisa 0402517015 RefleksiKurniah TunnisaNo ratings yet
- Rna Processing EukaryotesDocument33 pagesRna Processing EukaryotesNathanaelNo ratings yet
- DNA Replication: An OverviewDocument34 pagesDNA Replication: An OverviewSohini RoyNo ratings yet
- W4-1 Transcription StuDocument25 pagesW4-1 Transcription Stu23005965No ratings yet
- Sinh Tong Hop Rna - 2013Document41 pagesSinh Tong Hop Rna - 2013PhiPhiNo ratings yet
- Biological Sequence Determination: ProteinDocument68 pagesBiological Sequence Determination: ProteinamitNo ratings yet
- Science of Living System: Nihar Ranjan JanaDocument20 pagesScience of Living System: Nihar Ranjan JanaRajnandni SharmaNo ratings yet
- Lecturtry Operon 3Document4 pagesLecturtry Operon 3maxwell amponsahNo ratings yet
- An Introduction To Molecular BiologyDocument7 pagesAn Introduction To Molecular BiologyMaricon TubonNo ratings yet
- DNA Polymerase 2 TerminationDocument12 pagesDNA Polymerase 2 TerminationCemNo ratings yet
- MCQ MetabolicDocument5 pagesMCQ Metabolicghost beastNo ratings yet
- Science of Living System (BS20001) : - Soumya deDocument45 pagesScience of Living System (BS20001) : - Soumya deMayank PriayadarshiNo ratings yet
- Biology Lesson 1.1Document16 pagesBiology Lesson 1.1Crystal Joy BondadNo ratings yet
- DNA Replication: An OverviewDocument36 pagesDNA Replication: An OverviewRajnandni SharmaNo ratings yet
- Clase Biol Mol 11-05-10Document41 pagesClase Biol Mol 11-05-10Francisco RomeroNo ratings yet
- Transcription & RNA ProcessingDocument36 pagesTranscription & RNA ProcessingSubhajit AdakNo ratings yet
- BIOCORE 1 Midterm 2 Study GuideDocument44 pagesBIOCORE 1 Midterm 2 Study GuideWarren EnglishNo ratings yet
- Functional Activity of Commercial PrebioticsDocument6 pagesFunctional Activity of Commercial PrebioticshientricuongNo ratings yet
- Groove API 6ADocument3 pagesGroove API 6APedro de SousaNo ratings yet
- General-Mathematics ReviewerDocument2 pagesGeneral-Mathematics ReviewerEhranne EsperasNo ratings yet
- Chapter 50Document79 pagesChapter 50latrishahollmonNo ratings yet
- AP LAWCET 3 Years LLB 2015 Question Paper & Key PDFDocument1 pageAP LAWCET 3 Years LLB 2015 Question Paper & Key PDFpavaniNo ratings yet
- Tier1-Cvs, Cns and Respiratory Safety Pharmacology, Herg Assay Tier2-Gi, Renal and Other StudiesDocument30 pagesTier1-Cvs, Cns and Respiratory Safety Pharmacology, Herg Assay Tier2-Gi, Renal and Other StudiesCHETHAN K SNo ratings yet
- For Your Information Its MaggiDocument12 pagesFor Your Information Its MaggiSheetal SainiNo ratings yet
- Case Studies UNDP: GUASSA-MENZ COMMUNITY CONSERVATION AREADocument11 pagesCase Studies UNDP: GUASSA-MENZ COMMUNITY CONSERVATION AREAUNDP_EnvironmentNo ratings yet
- Mechanical Loads On Tubing Strings: © The Robert Gordon University 2006Document9 pagesMechanical Loads On Tubing Strings: © The Robert Gordon University 2006AlbertocNo ratings yet
- Polyfunctional MixturesDocument5 pagesPolyfunctional MixturesCarl Jerome CentenoNo ratings yet
- Brochure 3F Linda LED enDocument30 pagesBrochure 3F Linda LED enHarris SouglerisNo ratings yet
- DdarDocument300 pagesDdarbitok214No ratings yet
- CLE7 ThirdQuarter Module1Document10 pagesCLE7 ThirdQuarter Module1Roella Mae Tudias AlodNo ratings yet
- Westinghouse Lighting Remote Control of Outdoor Circuits Brochure 6-62Document12 pagesWestinghouse Lighting Remote Control of Outdoor Circuits Brochure 6-62Alan MastersNo ratings yet
- Class Prefixes Suffixes and Present Perfect Continuous 2021Document22 pagesClass Prefixes Suffixes and Present Perfect Continuous 2021Angelo Jorge ParedesNo ratings yet
- Gynae T and D Expl - 1Document44 pagesGynae T and D Expl - 1vivekanurag97No ratings yet
- Presentation 1 - Bio-BindersDocument15 pagesPresentation 1 - Bio-BindersidelgardNo ratings yet
- Delta Airlines and SummaryDocument39 pagesDelta Airlines and Summaryeissa alesayiNo ratings yet
- XBTG5230 PDFDocument112 pagesXBTG5230 PDFjcudrisNo ratings yet
- HOPE 3 Q1 Mod 4 VERSION 2.0Document15 pagesHOPE 3 Q1 Mod 4 VERSION 2.0Jhenery Henry IINo ratings yet
- Desquamative GingivitisDocument18 pagesDesquamative GingivitisSonia MehtaNo ratings yet