Professional Documents
Culture Documents
Abstract
Uploaded by
Amina InamOriginal Description:
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Abstract
Uploaded by
Amina InamCopyright:
Available Formats
PROTEIN ANALYSIS
GOVERNMENT COLLEGE UNIVERSITY, LAHORE.
DEPARTMENT OF CHEMISTRY
AMINA INAM
22-BH-BT-2017
“ROLE OF PROTEIN ANALYSIS IN
BIOTECHNOLOGY”
SIR-SHAH HUSSAIN
7-06-2019
ANALYTICAL CHEMISTRY Page 1
PROTEIN ANALYSIS
TABLE OF CONTENTS:-
* ABSTRACT
* KEY WORDS
* INTRODUCTION
* PROTEIN AND IT’S STRUCTURE
* TECHNIQUES OF PROTEIN ANALYSIS
* PROTEIN SEPARATION
* SDS-PAGE
* ISO-ELECTRIC FOCUSING
*PROTEIN SEQUENCING
* EDMAN DEGRADATION
* CHROMIC METHODS
* MASS SPECTROSCOPY
* ELISA
* WESTERN BLOTTING
* GEL ELECTROPHORESIS
*REFERNCES
ANALYTICAL CHEMISTRY Page 2
PROTEIN ANALYSIS
Abstract:
Being one of the impressing groups of macromolecules, proteins and their
application are crucial to understand the molecular logic of living cells. Being
composed of amino acids despite their similar structure, proteins are surrounded by
several other molecules of different chemical structures creating an overcrowded
environment that makes them different in their functions. Unraveling the
importance of protein folding, protein–protein interaction is critical to understand
cellular processes and rational drug design. A major breakthrough in protein
purification technologies has greatly changed the landscape of protein study
leading to a deeper understanding of their role in disease biochemical pathway and
also in drug metabolism. Since the first protein sequencing in 1953 by Frederic
Sanger, proteomics, which is the large-scale study of proteins, became an essential
tool in understanding the protein structures and functions. Furthermore, the
application of physicochemical measures for protein content has lit up comparative
and functional biology of proteins. This chapter is intended to give a rudimentary
understanding of the methods and technologies behind proteins and their
application.
ANALYTICAL CHEMISTRY Page 3
PROTEIN ANALYSIS
Keywords:-
*Proteins
*Protein Identification
*Protein Structural Analysis
*Protein purification
*ELISA
*Protein Quantitation
*Western blotting
ANALYTICAL CHEMISTRY Page 4
PROTEIN ANALYSIS
PROTEINS EXPLAINED:
Proteins, also known as polypeptides, are organic compounds made up of amino
acids. They are arranged in a linear chain and folded into a globular form. Proteins
are essential parts of organisms and they participate in virtually every process
within cells. Many proteins are enzymes that catalyse biochemical reactions and
are vital to metabolism. The size of a protein is an important physical characteristic
and scientists often use particle size analysers in their studies to discuss protein
size or molecular weight.
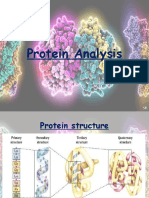
THE STRUCTURE OF PROTEINS:
Most proteins fold into unique three-dimensional structures. The shape that a
protein folds into naturally is known as its native conformation. While most
proteins can fold unassisted through the chemical properties of their amino acids,
others require the aid of molecular chaperones. There are four distinct aspects of a
protein’s structure:
Primary structure: the amino acid sequence
Secondary structure: regularly repeating local structures stabilised by hydrogen
bonds
Tertiary structure: the overall shape of a single protein molecule; the spatial
relationship of the secondary structures to one another
Quatemary structure: the structure formed by several protein molecules which
function as a single protein complex
PROTEIN ANALYSIS TECHNIQUES:
Proteins differ from each other according to the type, number and sequence of
amino acids that make up the polypeptide backbone. Hence, they have different
ANALYTICAL CHEMISTRY Page 5
PROTEIN ANALYSIS
molecular structures, nutritional attributes and physiochemical properties. There
are three major protein analysis techniques, protein separation, western blotting
and protein identification.
1. PROTEIN SEPARATION:
SDS-PAGE: This method separates proteins mainly on the basis of molecular
weight as opposed to charge or folding. It is a technique that is widely used in
biochemistry, forensics, genetics and molecular biology.
Isoelectric Focusing: In this method, different molecules are separated by their
electric charge differences. This technique is a type of zone electrophoresis that is
usually performed in a gel and takes advantage of the fact that a molecule’s charge
changes with the pH of its surroundings.
Chromatic Methods: There are two chromatic methods frequently used for
protein separation – high-performance liquid chromatography and thin-layer
chromatography. Both these methods are particularly useful adjuncts to gel-based
approaches. Although chromatography is a common technique in biochemistry
laboratories used for purification, identification and quantification of protein
mixtures, laser diffraction is traditionally used for pre-column size and
polydispersity management.
Two-dimensional Gel Electrophoresis: This is a powerful gel-based method
commonly used to analyse complex samples in the interest of characterising the
full range of proteins in the sample, not just a few specific proteins.
2. WESTERN BLOTTING
ANALYTICAL CHEMISTRY Page 6
PROTEIN ANALYSIS
The most common version of this method is immunoblotting. This technique is
used to detect specific proteins in a given sample of tissue homogenate or extract.
The sample of proteins is first electrophoresed by SDS-PAGE to separate the
proteins based on molecular weight. The proteins are then transferred to a
membrane where they are probed using antibodies specific to the target protein.
EXPLANATION:
Western Blot (WB) is a common method to detect and analyze proteins. It is built
on a technique that involves transferring, also known as blotting, proteins separated
by electrophoresis from the gel to a membrane where they can be visualized
specifically. The procedure was first described by H. Towbin et al in 1979
(Towbin, Staehelin, & Gordon, 1979) and two years later given its name by W.
Neal Burnette (Burnette, 1981). Towbin et al described electrophoretic transfer of
proteins from polyacrylamide gels to nitrocellulose sheets where the original gel
pattern was accurately obtained. The setup consists of a standard set of seven steps,
Figure .
Figure. The standard steps in Western Blotting.
Samples are prepared and loaded on to a gel and during the electrophoresis the
negatively charged proteins move toward the positively charged anode. In order to
further analyze the proteins, they are transferred onto a membrane in a procedure
called blotting. After the transfer, the membrane is blocked in order to prevent
unwanted membrane-protein interaction in the following steps. To visualize the
protein of interest the membrane is commonly first probed using a primary protein-
specific antibody followed by a labeled secondary antibody used for detection. An
image is taken of the membrane and the result is analyzed.
HOW PROCESS OCCUR?
By adding a separate marker solution to one of the wells in the gel, it is possible to
estimate the size of the protein in addition to the antibody interactions that are used
ANALYTICAL CHEMISTRY Page 7
PROTEIN ANALYSIS
to verify the specific protein. The separation on the gel is not only due to size but
also to some extent depending on the molecular charge, hydrophobic regions, and
degree of denaturation. The setup of the experiment can be varied in many ways to
best suit the specific inquiry. When analyzing the results, variations between lanes
regarding loading and transfer rates between blots, must be taken into
consideration. In addition, the non-linear relation of the generated signal across the
concentration range of the samples is also an aspect of consideration when
interpreting the results. The outcome of a WB experiment depends on three
important factors; the ability of the antibody to bind a specific protein, the strength
of the interaction, and the concentration of the protein of interest itself. Moreover,
the specificity of the binding to the target and a low cross reactivity are important
features as well. The result form the WB is not always easy to interpret as the size
of the protein may vary from the theoretical weight due to posttranslational
modifications, such as glycosylation, or interactions with other proteins. However,
WB is a very common method and almost all available commercial antibodies
have been validated .
Figure 5. Typical Western Blot result using HRP and a CCD camera.
ANALYTICAL CHEMISTRY Page 8
PROTEIN ANALYSIS
3. PROTEIN IDENTIFICATION
There are two methods that are commonly used to identify proteins.
Edman Degradation: Developed by Pehr Edman, this is a method of sequencing
amino acids in a peptide. Here, the amino-terminal residue is labeled and cleaved
from the peptide without disrupting the peptide bonds between other amino acid
residues.
EXPLANATION:
The Edman degradation is a very important reaction for protein sequencing,
because it allows the ordered amino acid composition of a protein to be discovered.
Automated Edman sequencers are now in widespread use, and are able to sequence
peptides up to approximately 50 amino acids long. A reaction scheme for
sequencing a protein by the Edman degradation follows; some of the steps are
elaborated on subsequently.
1. Break any disulfide bridges in the protein with a reducing agent like 2-
mercaptoethanol. A protecting group such as iodoacetic acid may be
necessary to prevent the bonds from re-forming.
2. Separate and purify the individual chains of the protein complex, if there are
more than one.
3. Determine the amino acid composition of each chain.
4. Determine the terminal amino acids of each chain.
5. Break each chain into fragments under 50 amino acids long.
6. Separate and purify the fragments.
7. Determine the sequence of each fragment.
8. Repeat with a different pattern of cleavage.
9. Construct the sequence of the overall protein.
Mass Spectrometry: An analytical technique that measures the mass-to-charge
ratio of charged particles, mass spectrometry is used for determining masses of
particles and the elemental composition of a sample of molecule as well as for
elucidating the chemical structure of molecules such as peptides.
PROTEIN SEQUENCING:
ANALYTICAL CHEMISTRY Page 9
PROTEIN ANALYSIS
Protein sequencing is the practical process of determining the amino acid
sequence of all or part of a protein or peptide. This may serve to identify the
protein or characterize its post-translational modifications. Typically, partial
sequencing of a protein provides sufficient information (one or more
sequence tags) to identify it with reference to databases of protein sequences
derived from the conceptual translation of genes.
The two major direct methods of protein sequencing are mass
spectrometry and Edman degradation using a protein
sequenator (sequencer). Mass spectrometry methods are now the most
widely used for protein sequencing and identification but Edman
degradation remains a valuable tool for characterizing a protein's N-
terminus.
It is often desirable to know the unordered amino acid composition of a protein
prior to attempting to find the ordered sequence, as this knowledge can be used to
facilitate the discovery of errors in the sequencing process or to distinguish
between ambiguous results. Knowledge of the frequency of certain amino acids
may also be used to choose which protease to use for digestion of the protein. The
misincorporation of low levels of non-standard amino acids (e.g. norleucine) into
proteins may also be determined. A generalized method often referred to as amino
acid analysis for determining amino acid frequency is as follows:
1. Hydrolyse a known quantity of protein into its constituent amino acids.
2. Separate and quantify the amino acids in some way.
Hydrolysis:
Hydrolysis is done by heating a sample of the protein in 6 M hydrochloric acid to
100–110 °C for 24 hours or longer. Proteins with many bulky hydrophobic groups
may require longer heating periods. However, these conditions are so vigorous that
some amino acids (serine, threonine, tyrosine, tryptophan, glutamine, and cysteine)
are degraded.
Separation and quantitation:
The amino acids can be separated by ion-exchange chromatography then
derivatized to facilitate their detection. More commonly, the amino acids are
derivatized then resolved by reversed phase HPLC.
An example of the ion-exchange chromatography is given by the NTRC using
sulfonated polystyrene as a matrix, adding the amino acids in acid solution and
passing a buffer of steadily increasing Ph through the column. Amino acids are
eluted when the pH reaches their respective isoelectric points.
ANALYTICAL CHEMISTRY Page 10
PROTEIN ANALYSIS
Gel electrophoresis is a method for separation and analysis of macromolecules
(DNA, RNA and proteins) and their fragments, based on their size and charge. It is
used in clinical chemistry to separate proteins by charge or size (IEF agarose,
essentially size independent) and in biochemistry and molecular biology to
separate a mixed population of DNA and RNA fragments by length, to estimate the
size of DNA and RNA fragments or to separate proteins by charge.
Nucleic acid molecules are separated by applying an electric field to move the
negatively charged molecules through a matrix of agarose or other substances.
Shorter molecules move faster and migrate farther than longer ones because shorter
molecules migrate more easily through the pores of the gel. This phenomenon is
called sieving. Proteins are separated by charge in agarose because the pores of the
gel are too large to sieve proteins. Gel electrophoresis can also be used for
separation of nanoparticles.
EXPLANATION:
Gel electrophoresis uses a gel as an anticonvective medium or sieving medium
during electrophoresis, the movement of a charged particle in an electrical field.
Gels suppress the thermal convection caused by application of the electric field,
and can also act as a sieving medium, retarding the passage of molecules; gels can
also simply serve to maintain the finished separation, so that a post electrophoresis
stain can be applied. DNA Gel electrophoresis is usually performed for analytical
purposes, often after amplification of DNA via polymerase chain reaction (PCR),
but may be used as a preparative technique prior to use of other methods such
as mass spectrometry, RFLP, PCR, cloning, DNA sequencing, or Southern
blotting for further characterization.
ANALYTICAL CHEMISTRY Page 11
PROTEIN ANALYSIS
Figure 3. The proteins in the gel are blotted to a membrane and the sample is
visualized through blocking, adding antibodies, and washing according to a certain
schedule.
HOW IT DIFFERNTIATED FROM OTHER TECHNIQUES?
After sample preparation the extract is ready to be loaded to separate the proteins
according to size by gel electrophoresis. An electric field is applied over the gel
that causes the charged molecules to move. In WB polyacrylamide gels are used
for protein separation and the method is therefore called polyacrylamide gel
electrophoresis (PAGE) when using native condition. For denaturing conditions,
sodium dodecyl sulfate (SDS) is added to the system and the method is therefore
called SDS-PAGE. The SDS binds to the protein and form a negatively charged
micelle around the protein regardless of inherent charge. The denaturing condition
dissolves the tridimensional structure of the proteins and the charge of the protein
becomes relative to its size resulting in separation of the proteins only by size.
When using native conditions the mobility is depending on both charge and
hydrodynamic size allowing detection of changes in charge due to chemical
degradation, conformational changes due to folding or unfolding, aggregation, or
other binding events.
TYPES OF GEL:
ANALYTICAL CHEMISTRY Page 12
PROTEIN ANALYSIS
The gel typically consists of two sections with different densities:
(i) a stacking gel
(ii) a separating gel.
The differences between the two sections are in pH and gel concentration. With
somewhat acidic pH and a lower concentration of acrylamide the stacking gel
separates the proteins poorly but allows them to form highly defined sharp bands
before they enter the separating gel. With more basic conditions and higher gel
concentration, the separating gel makes the proteins differentiate by size as smaller
proteins travel faster in the gel than bigger ones. Precast gels are convenient;
however, it is possible to cast them by hand. The gel is immersed in buffer and the
protein samples and markers are loaded to the wells in the gel. A voltage is applied
on the gel and the proteins will start to travel down the gel due to their negative
electrical charge. Selecting the proper voltage is important since too high voltage
will overheat the gel and maybe deform the bands.
ANALYTICAL CHEMISTRY Page 13
PROTEIN ANALYSIS
Figure 2. A typical gel immersed in buffer.
ELISA:
ELISA (enzyme-linked immunosorbent assay) is a plate-based assay technique
designed for detecting and quantifying substances such as peptides, proteins,
antibodies and hormones. Other names, such as enzyme immunoassay (EIA), are
also used to describe the same technology.
PROCESS OCCURING:
In an ELISA, an antigen must be immobilized on a solid surface and then
complexed with an antibody that is linked to an enzyme. Detection is accomplished
by assessing the conjugated enzyme activity via incubation with a substrate to
produce a measureable product. The most crucial element of the detection strategy
is a highly specific antibody-antigen interaction.
ELISAs are typically performed in 96-well (or 384-well) polystyrene plates, which
passively bind antibodies and proteins. It is this binding and immobilization of
reagents that makes ELISAs so easy to design and perform. Having the reactants of
the ELISA immobilized to the microplate surface makes it easy to separate bound
from non-bound material during the assay. This ability to wash away
nonspecifically bound materials makes the ELISA a powerful tool for measuring
specific analytes within a crude preparation.
A detection enzyme or other tag can be linked directly to the primary antibody or
introduced through a secondary antibody that recognizes the primary antibody. It
can also be linked to a protein such as streptavidin if the primary antibody is biotin
labeled. The most commonly used enzyme labels are horseradish peroxidase
(HRP) and alkaline phosphatase (AP). Other enzymes have been used as well, but
they have not gained widespread acceptance because of limited substrate options.
These include β-galactosidase, acetylcholinesterase and catalase. A large selection
of substrates is available for performing ELISA with an HRP or AP conjugate. The
choice of substrate depends upon the required assay sensitivity and the
instrumentation available for signal-detection (spectrophotometer, fluorometer or
luminometer).
ANALYTICAL CHEMISTRY Page 14
PROTEIN ANALYSIS
MODIFICATIONS:
ELISAs can be performed with a number of modifications to the basic procedure.
The key step, immobilization of the antigen of interest, can be accomplished by
direct adsorption to the assay plate or indirectly via a capture antibody that has
been attached to the plate. The antigen is then detected either directly (labeled
primary antibody) or indirectly (labeled secondary antibody). The most powerful
ELISA assay format is the sandwich assay. This type of capture assay is called a
“sandwich” assay because the analyte to be measured is bound between two
primary antibodies – the capture antibody and the detection antibody. The
sandwich format is used because it is sensitive and robust.
Diagram of common ELISA formats (direct vs. sandwich assays). In the assay,
the antigen of interest is immobilized by direct adsorption to the assay plate or by
first attaching a capture antibody to the plate surface. Detection of the antigen can
then be performed using an enzyme-conjugated primary antibody (direct detection)
or a matched set of unlabeled primary and conjugated secondary antibodies
(indirect detection).
Comparison of direct and indirect ELISA detection methods
Direct ELISA detection
Advantages Quick because only one antibody and fewer steps are used.
Cross-reactivity of secondary antibody is eliminated.
Disadvantages Immunoreactivity of the primary antibody might be adversely affected
by labeling with enzymes or tags.
ANALYTICAL CHEMISTRY Page 15
PROTEIN ANALYSIS
Labeling primary antibodies for each specific ELISA system is time-
consuming and expensive.
No flexibility in choice of primary antibody label from one experiment
to another.
Minimal signal amplification.
Indirect ELISA detection
Advantages A wide variety of labeled secondary antibodies are available
commercially.
Versatile because many primary antibodies can be made in one species
and the same labeled secondary antibody can be used for detection.
Maximum immunoreactivity of the primary antibody is retained
because it is not labeled.
Sensitivity is increased because each primary antibody contains several
epitopes that can be bound by the labeled secondary antibody, allowing
for signal amplification.
Different visualization markers can be used with the same primary
antibody.
Disadvantages Cross-reactivity might occur with the secondary antibody, resulting in
nonspecific signal.
An extra incubation step is required in the procedure.
ANALYTICAL CHEMISTRY Page 16
PROTEIN ANALYSIS
REFERNCES:
1. Voet, D. and J. G.Voet. 2011. Biochemistry, 4th Edition. John Wiley and Sons. Inc.,
New York.
2. David L. Nelson Michael M. Cox. 2008. Lehninger Analytical Chemistry, 5h
Edition. W.H. Freeman and Company, USA.
3. Campbell, M.K. and S.O. Farrell. 2006. 5th Edition.Biotechnolgy Techniques. Baba
BarkhaNath Printers, Delhi.
4. Nigam, A. and A. Ayyagari. 2008. Lab Manual in Biochemistry and
Biotechnology. McGraw-Hill and Companies, New Delhi.
ANALYTICAL CHEMISTRY Page 17
You might also like
- Assignment "Proteome Analysis and Its Applications" Paper Code: BIOTECH/04/SC/28 Paper Name: Genomics and ProteomicsDocument10 pagesAssignment "Proteome Analysis and Its Applications" Paper Code: BIOTECH/04/SC/28 Paper Name: Genomics and ProteomicsLalruatdiki CNo ratings yet
- Recombinant DNA Project Based Learning on Western BlottingDocument11 pagesRecombinant DNA Project Based Learning on Western Blottingvidushi srivastavaNo ratings yet
- 4 Protein 2022Document28 pages4 Protein 2022Hazem BaderNo ratings yet
- XII BiotechnologyDocument17 pagesXII BiotechnologyGuruKPONo ratings yet
- Protein Extraction and QuantificationDocument6 pagesProtein Extraction and QuantificationWNo ratings yet
- Enm 32 18 PDFDocument5 pagesEnm 32 18 PDFSofia andrea MezaNo ratings yet
- Unit 1: Structural GenomicsDocument4 pagesUnit 1: Structural GenomicsLavanya ReddyNo ratings yet
- MY Proteomics FinalDocument56 pagesMY Proteomics FinalSaradha PellatiNo ratings yet
- Metabolic Labeling of Proteins For Proteomics : Robert J. Beynon and Julie M. PrattDocument16 pagesMetabolic Labeling of Proteins For Proteomics : Robert J. Beynon and Julie M. PrattNidhi JaisNo ratings yet
- Methods of Proteins QuantificationDocument5 pagesMethods of Proteins QuantificationAnđela BošnjakNo ratings yet
- Labjournal1 For NursingDocument2 pagesLabjournal1 For NursingDanielle MatelNo ratings yet
- Proteins HeheheheDocument12 pagesProteins HeheheheErjel J. MalabananNo ratings yet
- Protein Molecular WeightDocument19 pagesProtein Molecular WeightMd Mostafa Kamal AbirNo ratings yet
- Iex of Biopharmaceuticals 1234Document15 pagesIex of Biopharmaceuticals 1234aa385864No ratings yet
- Purification of Protiens: Mohan CC M.SC, Biochemistry 1 SemesterDocument24 pagesPurification of Protiens: Mohan CC M.SC, Biochemistry 1 SemesterMelwin MejanNo ratings yet
- Introduction of ProteomicsDocument21 pagesIntroduction of ProteomicsMusfeera KhanNo ratings yet
- Characterizationofprotein 200505172732Document22 pagesCharacterizationofprotein 200505172732Yesica Marcelina SinagaNo ratings yet
- If Unit 4Document29 pagesIf Unit 4debajyoti pilaNo ratings yet
- Proteins: Nature's WorkhorsesDocument4 pagesProteins: Nature's WorkhorsesSatheesh KumarNo ratings yet
- Biochem Unit 5Document6 pagesBiochem Unit 5Elyon Jirehel AlvarezNo ratings yet
- s15 Miller Chap 3b LectureDocument25 pagess15 Miller Chap 3b LectureDorice Clement100% (1)
- Proteomics: by Hamsa Ehsan (16756) Maryam Saleem (16752) Namra Talib (16724)Document16 pagesProteomics: by Hamsa Ehsan (16756) Maryam Saleem (16752) Namra Talib (16724)Hamsa EhsanNo ratings yet
- Protein extraction and quantification from various organisms using Bradford AssayDocument5 pagesProtein extraction and quantification from various organisms using Bradford AssayBrandon Lam100% (1)
- Bio - Chap 4Document3 pagesBio - Chap 4iamNashix22No ratings yet
- Practical Applications of Proteomics-A Technique For Large-Scale Study of Proteins: An OverviewDocument4 pagesPractical Applications of Proteomics-A Technique For Large-Scale Study of Proteins: An OverviewVishwas gargNo ratings yet
- Immunoblotting Techniques: Principle, Methodology and ApplicationDocument18 pagesImmunoblotting Techniques: Principle, Methodology and Applicationnbanjare723No ratings yet
- Proteo MicsDocument25 pagesProteo MicsSaptarshi GhoshNo ratings yet
- Protein Microarray Naveed Up MushtaqDocument16 pagesProtein Microarray Naveed Up MushtaqAngumaniNo ratings yet
- A Short Review On Proteomics and Its ApplicationsDocument8 pagesA Short Review On Proteomics and Its ApplicationsMohamed HasanNo ratings yet
- Applications of ProteinDocument3 pagesApplications of ProteinMajid GhaffarNo ratings yet
- Methods in Molecular Biology, Vol.003 - New Protein TechniquesDocument524 pagesMethods in Molecular Biology, Vol.003 - New Protein TechniquesPablo HenrriquezNo ratings yet
- Chemical Proteomics in Drug Discovery and Target IDDocument3 pagesChemical Proteomics in Drug Discovery and Target IDFree Escort ServiceNo ratings yet
- Techniques For Protein PurificationDocument14 pagesTechniques For Protein PurificationTauqeer IqbalNo ratings yet
- Protein Purifi Cation: An Overview: Nikolaos E. LabrouDocument8 pagesProtein Purifi Cation: An Overview: Nikolaos E. LabrouSofia andrea MezaNo ratings yet
- Application of Proteomics for Discovery of Protein BiomarkersDocument9 pagesApplication of Proteomics for Discovery of Protein BiomarkersNidhi JaisNo ratings yet
- Subcellular Fractionation Methods and StrategiesDocument22 pagesSubcellular Fractionation Methods and StrategiesDaniel Juarez SerranoNo ratings yet
- 2023.10.10 MBG Proteins - Structure and FunctionDocument96 pages2023.10.10 MBG Proteins - Structure and Functionaida.mzreNo ratings yet
- Biochemistry Lecture Notes on Protein Structure and FunctionDocument15 pagesBiochemistry Lecture Notes on Protein Structure and FunctionRahaf Al-muhtasebNo ratings yet
- Gene Expression Studies: Transcriptomics and ProteomicsDocument69 pagesGene Expression Studies: Transcriptomics and ProteomicsZabrina Tan CuaNo ratings yet
- DocxDocument7 pagesDocxrobomunkey12345No ratings yet
- Garfin 1DE WebArticle9-07Document0 pagesGarfin 1DE WebArticle9-07Soma GhoshNo ratings yet
- How To Approach Bioinformatics Analysis in Protein Structure and FunctionDocument4 pagesHow To Approach Bioinformatics Analysis in Protein Structure and FunctionSuperSonic2422No ratings yet
- Assignment 4Document5 pagesAssignment 4Hafiz AhmadNo ratings yet
- Total Protein and Albumin DeterminationDocument12 pagesTotal Protein and Albumin DeterminationMaria ClaraNo ratings yet
- Lab Report Sds-Page WB - PT 1 (1-5)Document5 pagesLab Report Sds-Page WB - PT 1 (1-5)Ezad juferiNo ratings yet
- Faria & Halquist (2018) Internal Standards For Absolute Quantification of Large Moleculs by LCMSMSDocument25 pagesFaria & Halquist (2018) Internal Standards For Absolute Quantification of Large Moleculs by LCMSMStantry puspitasariNo ratings yet
- Proteomics Lab ReportDocument4 pagesProteomics Lab Reportnursah.aslanNo ratings yet
- Chapter 2 Protein SeparationDocument83 pagesChapter 2 Protein SeparationZul Adli SaribinNo ratings yet
- Proteomics - WikipediaDocument17 pagesProteomics - WikipediaP Bijoya SinghaNo ratings yet
- Protein Chips and Functional ProteomicsDocument9 pagesProtein Chips and Functional ProteomicsAmal ..No ratings yet
- Protein Micro ArrayDocument3 pagesProtein Micro ArrayBinil ManuelNo ratings yet
- Molecular Cell Biology Lodish 6th Edition Test BankDocument6 pagesMolecular Cell Biology Lodish 6th Edition Test BankCharles BlairNo ratings yet
- I. Title of Experiment: Determining The Value of Protein in ADocument29 pagesI. Title of Experiment: Determining The Value of Protein in AAnggraini Nugroho PNo ratings yet
- Multiprotein ComplexesDocument338 pagesMultiprotein Complexesgudangpdf02No ratings yet
- CH 5 Ky Thua Protein-Môn hóa sinh họcDocument83 pagesCH 5 Ky Thua Protein-Môn hóa sinh họcNam NguyenHoangNo ratings yet
- CH 5 Lecture SlidesDocument83 pagesCH 5 Lecture SlidesUyên Trần NhưNo ratings yet
- Proteomics Tools and Applications: A. EinsteinDocument62 pagesProteomics Tools and Applications: A. EinsteintejalNo ratings yet
- Proteomics analysis techniques and challengesDocument4 pagesProteomics analysis techniques and challengesDean PhoebeNo ratings yet
- Cell Biology Assays: ProteinsFrom EverandCell Biology Assays: ProteinsFanny JaulinNo ratings yet
- 2022 PCRDocument7 pages2022 PCRb409110037No ratings yet
- Agarose Gel Electrophoresis of DNA PDFDocument5 pagesAgarose Gel Electrophoresis of DNA PDFBiologistAhmedNo ratings yet
- Unicel Dxi Reference Manual 1.2: Assay TechnologyDocument1 pageUnicel Dxi Reference Manual 1.2: Assay TechnologysantoshNo ratings yet
- Promo Pierce h2 - Promo VenditaDocument2 pagesPromo Pierce h2 - Promo VenditalacewingNo ratings yet
- Western Blot Membrane Stripping For Restaining ProtocolDocument2 pagesWestern Blot Membrane Stripping For Restaining ProtocolDouglas SantosNo ratings yet
- History of Northern BlottingDocument5 pagesHistory of Northern BlottingNur SyahirahNo ratings yet
- BIO 103 General Biology: Agarose Gel ElectrophoresisDocument13 pagesBIO 103 General Biology: Agarose Gel ElectrophoresisseydatkrNo ratings yet
- Gel ElectroiphoresisDocument22 pagesGel ElectroiphoresisAqsa MazharNo ratings yet
- MagCore Kit 601 DMDocument2 pagesMagCore Kit 601 DMBogdan NeamtuNo ratings yet
- Amina Bashir 20700257 Agarose Gel ElectrophoresisDocument5 pagesAmina Bashir 20700257 Agarose Gel Electrophoresishamza vardarNo ratings yet
- Blotting Techniques2 PDFDocument3 pagesBlotting Techniques2 PDFLolaNo ratings yet
- Genei: Western Blotting Teaching Kit ManualDocument10 pagesGenei: Western Blotting Teaching Kit ManualSoma GhoshNo ratings yet
- Experiment 2:dna TechnologyDocument6 pagesExperiment 2:dna TechnologycheckerskNo ratings yet
- ICoBioSE Real-Time QPCR Application Workshop - Gene Expression AnalysisDocument10 pagesICoBioSE Real-Time QPCR Application Workshop - Gene Expression Analysisafifatul achyarNo ratings yet
- 1Kbp DNA Ladder Marker Ready-to-UseDocument2 pages1Kbp DNA Ladder Marker Ready-to-UsexprakashNo ratings yet
- DNA Gel ElectrophoresisDocument5 pagesDNA Gel ElectrophoresisTok WanNo ratings yet
- Agarose Gel ElectrophoresisDocument6 pagesAgarose Gel ElectrophoresisJêyà BharathìNo ratings yet
- Package: Catalog No. SizeDocument3 pagesPackage: Catalog No. SizeHairul IslamNo ratings yet
- AbD Serotec Antibodies France - rEDDocument1,323 pagesAbD Serotec Antibodies France - rEDrdLuis1No ratings yet
- Gel filtration separates molecules by sizeDocument17 pagesGel filtration separates molecules by sizeANU CHOUDHARYNo ratings yet
- Protocolo Gelred PDFDocument2 pagesProtocolo Gelred PDFRodrigo UchyamaNo ratings yet
- SDS-PAGE Guide: Separate Proteins by Size & ChargeDocument39 pagesSDS-PAGE Guide: Separate Proteins by Size & ChargeSnehalphirke100% (1)
- Agarose Gel ElectrophoresisDocument4 pagesAgarose Gel ElectrophoresisShailendra YadavNo ratings yet
- MiniPCR P51 Intro To QPCR Lab Classroom Slides v1 4Document32 pagesMiniPCR P51 Intro To QPCR Lab Classroom Slides v1 4Areej ArshadNo ratings yet
- Identification of Unknown PlasmidDocument9 pagesIdentification of Unknown Plasmidapi-233148262No ratings yet
- Sds Page ProtocolDocument2 pagesSds Page Protocolbiosa45No ratings yet
- Gel Electrophoresis - Wikipedia, The Free EncyclopediaDocument10 pagesGel Electrophoresis - Wikipedia, The Free EncyclopediaShailendra YadavNo ratings yet
- GFP TrapDocument4 pagesGFP TrapAllele BiotechnologyNo ratings yet
- Software errors and their effectsDocument157 pagesSoftware errors and their effectsОлександрNo ratings yet
- Gel Electrophoresis: Separating Molecules by Size & ChargeDocument5 pagesGel Electrophoresis: Separating Molecules by Size & ChargeSai SridharNo ratings yet