Professional Documents
Culture Documents
TenRules PDF
TenRules PDF
Uploaded by
HeitorOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
TenRules PDF
TenRules PDF
Uploaded by
HeitorCopyright:
Available Formats
Editorial
Ten Simple Rules for Reproducible Computational
Research
Geir Kjetil Sandve1,2*, Anton Nekrutenko3, James Taylor4, Eivind Hovig1,5,6
1 Department of Informatics, University of Oslo, Blindern, Oslo, Norway, 2 Centre for Cancer Biomedicine, University of Oslo, Blindern, Oslo, Norway, 3 Department of
Biochemistry and Molecular Biology and The Huck Institutes for the Life Sciences, Penn State University, University Park, Pennsylvania, United States of America,
4 Department of Biology and Department of Mathematics and Computer Science, Emory University, Atlanta, Georgia, United States of America, 5 Department of Tumor
Biology, Institute for Cancer Research, The Norwegian Radium Hospital, Oslo University Hospital, Montebello, Oslo, Norway, 6 Institute for Medical Informatics, The
Norwegian Radium Hospital, Oslo University Hospital, Montebello, Oslo, Norway
Replication is the cornerstone of a We further note that reproducibility is than to do it while underway). We believe
cumulative science [1]. However, new just as much about the habits that ensure that the rewards of reproducibility will
tools and technologies, massive amounts reproducible research as the technologies compensate for the risk of having spent
of data, interdisciplinary approaches, and that can make these processes efficient and valuable time developing an annotated
the complexity of the questions being realistic. Each of the following ten rules catalog of analyses that turned out as blind
asked are complicating replication efforts, captures a specific aspect of reproducibil- alleys.
as are increased pressures on scientists to ity, and discusses what is needed in terms As a minimal requirement, you should
advance their research [2]. As full replica- of information handling and tracking of at least be able to reproduce the results
tion of studies on independently collected procedures. If you are taking a bare-bones yourself. This would satisfy the most basic
data is often not feasible, there has recently approach to bioinformatics analysis, i.e., requirements of sound research, allowing
been a call for reproducible research as an running various custom scripts from the any substantial future questioning of the
attainable minimum standard for assessing command line, you will probably need to research to be met with a precise expla-
the value of scientific claims [3]. This handle each rule explicitly. If you are nation. Although it may sound like a very
requires that papers in experimental instead performing your analyses through weak requirement, even this level of
science describe the results and provide a an integrated framework (such as Gene- reproducibility will often require a certain
sufficiently clear protocol to allow success- Pattern [10], Galaxy [11], LONI pipeline level of care in order to be met. There will
ful repetition and extension of analyses [12], or Taverna [13]), the system may for a given analysis be an exponential
based on original data [4]. already provide full or partial support for number of possible combinations of soft-
The importance of replication and most of the rules. What is needed on your ware versions, parameter values, pre-
reproducibility has recently been exempli- part is then merely the knowledge of how processing steps, and so on, meaning that
fied through studies showing that scientific to exploit these existing possibilities. a failure to take notes may make exact
papers commonly leave out experimental In a pragmatic setting, with publication reproduction essentially impossible.
details essential for reproduction [5], pressure and deadlines, one may face the With this basic level of reproducibility in
studies showing difficulties with replicating need to make a trade-off between the place, there is much more that can be
published experimental results [6], an ideals of reproducibility and the need to wished for. An obvious extension is to go
increase in retracted papers [7], and get the research out while it is still relevant. from a level where you can reproduce
through a high number of failing clinical This trade-off becomes more important results in case of a critical situation to a
trials [8,9]. This has led to discussions on when considering that a large part of the level where you can practically and
how individual researchers, institutions, analyses being tried out never end up routinely reuse your previous work and
funding bodies, and journals can establish yielding any results. However, frequently increase your productivity. A second
routines that increase transparency and one will, with the wisdom of hindsight, extension is to ensure that peers have a
reproducibility. In order to foster such contemplate the missed opportunity to practical possibility of reproducing your
aspects, it has been suggested that the ensure reproducibility, as it may already results, which can lead to increased trust
scientific community needs to develop a be too late to take the necessary notes from in, interest for, and citations of your work
‘‘culture of reproducibility’’ for computa- memory (or at least much more difficult [6,14].
tional science, and to require it for
published claims [3].
Citation: Sandve GK, Nekrutenko A, Taylor J, Hovig E (2013) Ten Simple Rules for Reproducible Computational
We want to emphasize that reproduc- Research. PLoS Comput Biol 9(10): e1003285. doi:10.1371/journal.pcbi.1003285
ibility is not only a moral responsibility
Editor: Philip E. Bourne, University of California San Diego, United States of America
with respect to the scientific field, but that
Published October 24, 2013
a lack of reproducibility can also be a
burden for you as an individual research- Copyright: ß 2013 Sandve et al. This is an open-access article distributed under the terms of the Creative
Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium,
er. As an example, a good practice of provided the original author and source are credited.
reproducibility is necessary in order to
Funding: The authors’ laboratories are supported by US National Institutes of Health grants HG005133,
allow previously developed methodology HG004909, and HG006620 and US National Science Foundation grant DBI 0850103. Additional funding is
to be effectively applied on new data, or to provided, in part, by the Huck Institutes for the Life Sciences at Penn State, the Institute for Cyberscience at
allow reuse of code and results for new Penn State, and a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds. The
funders had no role in the preparation of the manuscript.
projects. In other words, good habits of
reproducibility may actually turn out to be Competing Interests: The authors have declared that no competing interests exist.
a time-saver in the longer run. * E-mail: geirksa@ifi.uio.no
PLOS Computational Biology | www.ploscompbiol.org 1 October 2013 | Volume 9 | Issue 10 | e1003285
We here present ten simple rules for files can usually be replaced by the use of hopeless task. This can cast doubt on
reproducibility of computational research. standard UNIX commands or small previous results, as it may be impossible to
These rules can be at your disposal for custom scripts. If working with integrated know if they were partly the result of a bug
whenever you want to make your research frameworks, there will typically be a quite or otherwise unfortunate behavior.
more accessible—be it for peers or for rich collection of components for data The standard solution to track evolution
your future self. manipulation. As an example, manual of code is to use a version control system
tweaking of data files to attain format [15], such as Subversion, Git, or Mercu-
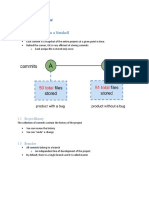
Rule 1: For Every Result, Keep compatibility should be replaced by for- rial. These systems are relatively easy to set
Track of How It Was Produced mat converters that can be reenacted and up and use, and may be used to system-
included into executable workflows. Other atically store the state of the code through-
Whenever a result may be of potential manual operations like the use of copy and out development at any desired time
interest, keep track of how it was pro- paste between documents should also be granularity.
duced. When doing this, one will frequent- avoided. If manual operations cannot be As a minimum, you should archive
ly find that getting from raw data to the avoided, you should as a minimum note copies of your scripts from time to time,
final result involves many interrelated down which data files were modified or so that you keep a rough record of the
steps (single commands, scripts, programs). moved, and for what purpose. various states the code has taken during
We refer to such a sequence of steps, development.
whether it is automated or performed Rule 3: Archive the Exact
manually, as an analysis workflow. While Versions of All External Rule 5: Record All Intermediate
the essential part of an analysis is often Results, When Possible in
Programs Used
represented by only one of the steps, the
Standardized Formats
full sequence of pre- and post-processing In order to exactly reproduce a given
steps are often critical in order to reach the result, it may be necessary to use programs In principle, as long as the full process
achieved result. For every involved step, in the exact versions used originally. Also, used to produce a given result is tracked,
you should ensure that every detail that as both input and output formats may all intermediate data can also be regener-
may influence the execution of the step is change between versions, a newer version ated. In practice, having easily accessible
recorded. If the step is performed by a of a program may not even run without intermediate results may be of great value.
computer program, the critical details modifying its inputs. Even having noted Quickly browsing through intermediate
include the name and version of the which version was used of a given results can reveal discrepancies toward
program, as well as the exact parameters program, it is not always trivial to get what is assumed, and can in this way
and inputs that were used. hold of a program in anything but the uncover bugs or faulty interpretations that
Although manually noting the precise current version. Archiving the exact ver- are not apparent in the final results.
sequence of steps taken allows for an sions of programs actually used may thus Secondly, it more directly reveals conse-
analysis to be reproduced, the documen- save a lot of hassle at later stages. In some quences of alternative programs and
tation can easily get out of sync with how cases, all that is needed is to store a single parameter choices at individual steps.
the analysis was really performed in its executable or source code file. In other Thirdly, when the full process is not
final version. By instead specifying the full cases, a given program may again have readily executable, it allows parts of the
analysis workflow in a form that allows for specific requirements to other installed process to be rerun. Fourthly, when
direct execution, one can ensure that the programs/packages, or dependencies to reproducing results, it allows any experi-
specification matches the analysis that was specific operating system components. To enced inconsistencies to be tracked to the
(subsequently) performed, and that the ensure future availability, the only viable steps where the problems arise. Fifth, it
analysis can be reproduced by yourself or solution may then be to store a full virtual allows critical examination of the full
others in an automated way. Such execut- machine image of the operating system process behind a result, without the need
able descriptions [10] might come in the and program. As a minimum, you should to have all executables operational. When
form of simple shell scripts or makefiles note the exact names and versions of the possible, store such intermediate results in
[15,16] at the command line, or in the main programs you use. standardized formats. As a minimum,
form of stored workflows in a workflow archive any intermediate result files that
management system [10,11,13,17,18]. Rule 4: Version Control All are produced when running an analysis (as
As a minimum, you should at least Custom Scripts long as the required storage space is not
record sufficient details on programs, prohibitive).
parameters, and manual procedures to Even the slightest change to a computer
allow yourself, in a year or so, to program can have large intended or Rule 6: For Analyses That
approximately reproduce the results. unintended consequences. When a contin- Include Randomness, Note
ually developed piece of code (typically a
Underlying Random Seeds
Rule 2: Avoid Manual Data small script) has been used to generate a
Manipulation Steps certain result, only that exact state of the Many analyses and predictions include
script may be able to produce that exact some element of randomness, meaning the
Whenever possible, rely on the execu- output, even given the same input data same program will typically give slightly
tion of programs instead of manual and parameters. As also discussed for rules different results every time it is executed
procedures to modify data. Such manual 3 and 6, exact reproduction of results may (even when receiving identical inputs and
procedures are not only inefficient and in certain situations be essential. If com- parameters). However, given the same
error-prone, they are also difficult to puter code is not systematically archived initial seed, all random numbers used in
reproduce. If working at the UNIX along its evolution, backtracking to a code an analysis will be equal, thus giving
command line, manual modification of state that gave a certain result may be a identical results every time it is run. There
PLOS Computational Biology | www.ploscompbiol.org 2 October 2013 | Volume 9 | Issue 10 | e1003285
is a large difference between observing highly summarized data. For instance, as it may be hard to locate the exact
that a result has been reproduced exactly each value along a curve may in turn result underlying and supporting the
or only approximately. While achieving represent averages from an underlying statement from a large pool of different
equal results is a strong indication that a distribution. In order to validate and analyses with various versions.
procedure has been reproduced exactly, it fully understand the main result, it is To allow efficient retrieval of details
is often hard to conclude anything when often useful to inspect the detailed values behind textual statements, we suggest
achieving only approximately equal re- underlying the summaries. A common that statements are connected to under-
sults. For analyses that involve random but impractical way of doing this is to lying results already from the time the
numbers, this means that the random seed incorporate various debug outputs in the statements are initially formulated (for
should be recorded. This allows results to source code of scripts and programs. instance in notes or emails). Such a
be reproduced exactly by providing the When the storage context allows, it is connection can for instance be a simple
same seed to the random number gener- better to simply incorporate permanent file path to detailed results, or the ID of a
ator in future runs. As a minimum, you output of all underlying data when a result in an analysis framework, included
should note which analysis steps involve main result is generated, using a system- within the text itself. For an even tighter
randomness, so that a certain level of atic naming convention to allow the full integration, there are tools available to
discrepancy can be anticipated when data underlying a given summarized help integrate reproducible analyses di-
reproducing the results. value to be easily found. We find rectly into textual documents, such as
hypertext (i.e., html file output) to be Sweave [19], the GenePattern Word
Rule 7: Always Store Raw Data particularly useful for this purpose. This add-in [4], and Galaxy Pages [20]. These
behind Plots allows summarized results to be generat- solutions can also subsequently be used
ed along with links that can be very in connection with publications, as dis-
From the time a figure is first generated conveniently followed (by simply click- cussed in the next rule.
to it being part of a published article, it is ing) to the full data underlying each
often modified several times. In some As a minimum, you should provide
summarized value. When working with
cases, such modifications are merely visual enough details along with your textual
summarized results, you should as a
adjustments to improve readability, or to interpretations so as to allow the exact
minimum at least once generate, inspect,
ensure visual consistency between figures. underlying results, or at least some
and validate the detailed values underly-
If raw data behind figures are stored in a related results, to be tracked down in
ing the summaries.
systematic manner, so as to allow raw data the future.
for a given figure to be easily retrieved, Rule 9: Connect Textual
one can simply modify the plotting Rule 10: Provide Public Access
Statements to Underlying
procedure, instead of having to redo the to Scripts, Runs, and Results
whole analysis. An additional advantage of Results
this is that if one really wants to read fine Last, but not least, all input data,
Throughout a typical research project, a
values in a figure, one can consult the raw scripts, versions, parameters, and inter-
range of different analyses are tried and
numbers. In cases where plotting involves mediate results should be made publicly
interpretation of the results made. Al-
more than a direct visualization of under- and easily accessible. Various solutions
though the results of analyses and their
lying numbers, it can be useful to store have now become available to make data
corresponding textual interpretations are
both the underlying data and the pro- clearly interconnected at the conceptual sharing more convenient, standardized,
cessed values that are directly visualized. level, they tend to live quite separate lives and accessible in particular domains,
An example of this is the plotting of in their representations: results usually such as for gene expression data [21–
histograms, where both the values before live on a data area on a server or 23]. Most journals allow articles to be
binning (original data) and the counts per personal computer, while interpretations supplemented with online material, and
bin (heights of visualized bars) could be live in text documents in the form of some journals have initiated further
stored. When plotting is performed using a personal notes or emails to collaborators. efforts for making data and code more
command-based system like R, it is Such textual interpretations are not integrated with publications [3,24]. As a
convenient to also store the code used to generally mere shadows of the results— minimum, you should submit the main
make the plot. One can then apply slight they often involve viewing the results in data and source code as supplementary
modifications to these commands, instead light of other theories and results. As material, and be prepared to respond to
of having to specify the plot from scratch. such, they carry extra information, while any requests for further data or method-
As a minimum, one should note which at the same time having their necessary ology details by peers.
data formed the basis of a given plot and support in a given result. Making reproducibility of your work
how this data could be reconstructed. If you want to reevaluate your previ- by peers a realistic possibility sends a
ous interpretations, or allow peers to strong signal of quality, trustworthiness,
Rule 8: Generate Hierarchical make their own assessment of claims you and transparency. This could increase
Analysis Output, Allowing make in a scientific paper, you will have the quality and speed of the reviewing
Layers of Increasing Detail to Be to connect a given textual statement process on your work, the chances of
(interpretation, claim, conclusion) to the your work getting published, and the
Inspected
precise results underlying the statement. chances of your work being taken further
The final results that make it to an Making this connection when it is and cited by other researchers after
article, be it plots or tables, often represent needed may be difficult and error-prone, publication [25].
PLOS Computational Biology | www.ploscompbiol.org 3 October 2013 | Volume 9 | Issue 10 | e1003285
References
1. Crocker J, Cooper ML (2011) Addressing scien- 11. Giardine B, Riemer C, Hardison RC, Burhans R, systems. Scientific Programming Journal 13:
tific fraud. Science 334: 1182. Elnitski L, et al. (2005) Galaxy: a platform for 219–237.
2. Jasny BR, Chin G, Chong L, Vignieri S (2011) interactive large-scale genome analysis. Genome 19. Leisch F (2002) Sweave: dynamic generation of
Data replication & reproducibility. Again, and Res 15: 1451–1455. statistical reports using literate data analysis. In:
again, and again…. Introduction. Science 334: 12. Rex DE, Ma JQ, Toga AW (2003) The LONI Härdle W, Rönz B, editors. Compstat: proceed-
1225. Pipeline Processing Environment. Neuroimage ings in computational statistics. Heidelberg,
3. Peng RD (2011) Reproducible research in 19: 1033–1048. Germany: Physika Verlag. pp. 575–580.
computational science. Science 334: 1226–1227. 13. Oinn T, Addis M, Ferris J, Marvin D, Senger M, 20. Goecks J, Nekrutenko A, Taylor J (2010) Galaxy:
4. Mesirov JP (2010) Computer science. Accessible et al. (2004) Taverna: a tool for the composition a comprehensive approach for supporting acces-
reproducible research. Science 327: 415–416. and enactment of bioinformatics workflows. sible, reproducible, and transparent computation-
5. Nekrutenko A, Taylor J (2012) Next-generation Bioinformatics 20: 3045–3054. al research in the life sciences. Genome Biol 11:
sequencing data interpretation: enhancing repro- 14. Piwowar HA, Day RS, Fridsma DB (2007) R86.
ducibility and accessibility. Nat Rev Genet 13: Sharing detailed research data is associated with 21. Brazma A, Hingamp P, Quackenbush J, Sherlock
increased citation rate. PLoS ONE 2: e308. G, Spellman P, et al. (2001) Minimum informa-
667–672.
tion about a microarray experiment (MIAME)-
6. Ioannidis JP, Allison DB, Ball CA, Coulibaly I, doi:10.1371/journal.pone.0000308.
toward standards for microarray data. Nat Genet
Cui X, et al. (2009) Repeatability of published 15. Heroux MA, Willenbring JM (2009) Barely
29: 365–371.
microarray gene expression analyses. Nat Genet sufficient software engineering: 10 practices to
22. Brazma A, Parkinson H, Sarkans U, Shojatalab
41: 149–155. improve your cse software. In: 2009 ICSE
M, Vilo J, et al. (2003) ArrayExpress–a public
7. Steen RG (2011) Retractions in the scientific Workshop on Software Engineering for Compu- repository for microarray gene expression data at
literature: is the incidence of research fraud tational Science and Engineering. pp. 15–21. the EBI. Nucleic Acids Res 31: 68–71.
increasing? J Med Ethics 37: 249–253. 16. Schwab M, Karrenbach M, Claerbout J (2000) 23. Edgar R, Domrachev M, Lash AE (2002) Gene
8. Prinz F, Schlange T, Asadullah K (2011) Believe Making scientific computations reproducible. Expression Omnibus: NCBI gene expression and
it or not: how much can we rely on published data Comput Sci Eng 2: 61–67. hybridization array data repository. Nucleic Acids
on potential drug targets? Nat Rev Drug Discov 17. Goble CA, Bhagat J, Aleksejevs S, Cruickshank Res 30: 207–210.
10: 712. D, Michaelides D, et al. (2010) myExperiment: a 24. Sneddon TP, Li P, Edmunds SC (2012) GigaDB:
9. Begley CG, Ellis LM (2012) Drug development: repository and social network for the sharing of announcing the GigaScience database. Giga-
raise standards for preclinical cancer research. bioinformatics workflows. Nucleic Acids Res 38: science 1: 11.
Nature 483: 531–533. W677–682. 25. Prlić A, Procter JB (2012) Ten simple rules for the
10. Reich M, Liefeld T, Gould J, Lerner J, Tamayo 18. Deelman E, Singh G, Su M-H, Blythe J, Gil Y, et open development of scientific software. PLoS
P, et al. (2006) GenePattern 2.0. Nat Genet 38: al. (2005) Pegasus: a framework for mapping Comput Biol 8: e1002802. doi:10.1371/journal.-
500–501. complex scientific workflows onto distributed pcbi.1002802.
PLOS Computational Biology | www.ploscompbiol.org 4 October 2013 | Volume 9 | Issue 10 | e1003285
You might also like
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (122)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (266)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (590)
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (842)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5807)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2259)
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (401)
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (897)
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1091)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (346)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (74)
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- Data Integration Life Cycle Management With SSIS - A Short Introduction by ExampleDocument188 pagesData Integration Life Cycle Management With SSIS - A Short Introduction by ExampleAndreasNo ratings yet
- Git TutorialDocument35 pagesGit TutorialRoberto Martinez100% (1)
- Software Engineering - Technical Publication - 230625 - 090636Document281 pagesSoftware Engineering - Technical Publication - 230625 - 090636YZ-AMVNo ratings yet
- FactoryTalk AssetCentre Basic Configuration LabDocument89 pagesFactoryTalk AssetCentre Basic Configuration LabSteveNo ratings yet
- RT Technologies +91-9019995361: Purushotham ReddyDocument17 pagesRT Technologies +91-9019995361: Purushotham ReddySai Ganesh Reddy Kasireddy100% (1)
- Arc 1 Mod 1 - Architect Overview - v2.1Document73 pagesArc 1 Mod 1 - Architect Overview - v2.1tharun saiNo ratings yet
- Seeds Ii R1Document42 pagesSeeds Ii R1alnemerjassem100% (1)
- Software Configuration ManagementDocument12 pagesSoftware Configuration ManagementRishabh Makhija50% (10)
- Quest StatDocument2 pagesQuest Statsrpothul100% (1)
- Latex Thesis TemplateDocument7 pagesLatex Thesis Templatejennacaldejonanchorage100% (2)
- B.Naveen Reddy Email: Mobile No: +91-8919 523 901 Career ObjectiveDocument3 pagesB.Naveen Reddy Email: Mobile No: +91-8919 523 901 Career Objectiveakkati123No ratings yet
- Database Metadata: Note This Article Refers To An Example Sharepoint Site Created by Adventure Works, A FictitiousDocument4 pagesDatabase Metadata: Note This Article Refers To An Example Sharepoint Site Created by Adventure Works, A FictitiousLuky KurniawanNo ratings yet
- Tutorial Windev 20Document453 pagesTutorial Windev 20Damasio Mancuello RodasNo ratings yet
- 1 Devops and Git in A NutshellDocument30 pages1 Devops and Git in A NutshellsadoNo ratings yet
- Shivashankar (4 3)Document3 pagesShivashankar (4 3)k maheshNo ratings yet
- Techtalk Devops Slide DeckDocument65 pagesTechtalk Devops Slide DeckCaricarisoftNo ratings yet
- Github Workflow: According To GoogleDocument16 pagesGithub Workflow: According To GooglebengaltigerNo ratings yet
- Project Tittle: Wolkite University Medical Centre Management SystemDocument41 pagesProject Tittle: Wolkite University Medical Centre Management SystemyonasNo ratings yet
- Git Cheatsheet: Inspect Repository Edit Repository Create A RepositoryDocument2 pagesGit Cheatsheet: Inspect Repository Edit Repository Create A RepositoryKiril IvanovNo ratings yet
- Software Configuration ManagementDocument24 pagesSoftware Configuration ManagementHaveit12No ratings yet
- Should Cost Modelling Guidance Note May 2021Document24 pagesShould Cost Modelling Guidance Note May 2021Kishora BhatNo ratings yet
- Heinzelman, Matt - Software Quality Assurance (Techniques and Tools)Document10 pagesHeinzelman, Matt - Software Quality Assurance (Techniques and Tools)syrexNo ratings yet
- Plastic-Gui EsDocument126 pagesPlastic-Gui EsLucky HunterNo ratings yet
- Devops RecordDocument109 pagesDevops RecordPriya NeelamNo ratings yet
- GitDocument2 pagesGitrajehshsjNo ratings yet
- PHD Thesis Latex TemplateDocument6 pagesPHD Thesis Latex Templateaflohdtogglebv100% (2)
- Perforce p4vDocument2 pagesPerforce p4vSravanthi MaddineniNo ratings yet
- OpenText StreamServe 5.6.2 Control Center User GuideDocument228 pagesOpenText StreamServe 5.6.2 Control Center User GuideDennis van den BergNo ratings yet
- Jenkins PipelineDocument5 pagesJenkins Pipelinejyothirmai ManikyamNo ratings yet
- Top Answers To Git Interview QuestionsDocument2 pagesTop Answers To Git Interview QuestionsejazalmNo ratings yet