Professional Documents
Culture Documents
Supporting Information:: The More Frequent Snvs in The Genome of Wuhan/Ivdchb05/2019 - Epi - Isl - 402121 Are
Uploaded by
Shayan MuazzamOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Supporting Information:: The More Frequent Snvs in The Genome of Wuhan/Ivdchb05/2019 - Epi - Isl - 402121 Are
Uploaded by
Shayan MuazzamCopyright:
Available Formats
1 Supporting Information:
2
3
4
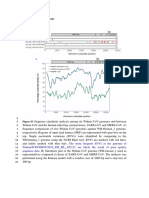
5 Figure S1 Sequence similarity analysis among six Wuhan CoV genomes and between
6 Wuhan CoV and the human-infecting coronaviruses, SARS-CoV and MERS-CoV. A)
7 Sequence comparison of five Wuhan CoV genomes against WH-Human_1 genome,
8 respectively. Regions of open read frames (ORF) are represented with gray box on the
9 top. Single nucleotide variations (SNVs) were identified by comparing to the
10 WH-human_1 genome using the NCBI Blast tool. SNVs are marked with red lines,
11 and indels marked with blue bars. The more frequent SNVs in the genome of
12 Wuhan/IVDCHB05/2019_EPI_ISL_402121 are likely the result of poor quality
13 sequence data. B) Similarity plot of the Wuhan CoV genome (WH-human_1 used as
14 representative) compared to those of SARS-CoV and MERS-CoV. The analysis was
15 performed using the Kimura model with a window size of 1000 bp and a step size of
16 200 bp.
You might also like
- 2020 1637 Moesm1 Esm PDFDocument1 page2020 1637 Moesm1 Esm PDFShayan MuazzamNo ratings yet
- Whole Genome Sequencing Is More PowerfulDocument6 pagesWhole Genome Sequencing Is More PowerfulluismoralesmNo ratings yet
- Diagnostic Usefulness of Dermoscopy in Differentiating Lichen Aureus From Nummular EczemaDocument5 pagesDiagnostic Usefulness of Dermoscopy in Differentiating Lichen Aureus From Nummular EczemaFebrian AlfaroNo ratings yet
- Wiszniewska 2014 Combined Array CGH Plus SNP GenomeDocument9 pagesWiszniewska 2014 Combined Array CGH Plus SNP GenomeSwathi sampathkumarNo ratings yet
- Copy Number Variation: Tie-Lin Yang, Yan Guo, Christopher J. Papasian and Hong-Wen DengDocument1 pageCopy Number Variation: Tie-Lin Yang, Yan Guo, Christopher J. Papasian and Hong-Wen DengBenyam ZenebeNo ratings yet
- 2011.diagnostic Implications of Excessive Homozygosity Detected by SNP Based MicroarrayDocument19 pages2011.diagnostic Implications of Excessive Homozygosity Detected by SNP Based Microarrayrpina.genNo ratings yet
- Li - Som Supplement PaperDocument149 pagesLi - Som Supplement PaperInambioinfoNo ratings yet
- A Sars Like Cluster of Circulating Bat Coronaviruses Shows Potential For Human EmergenceDocument17 pagesA Sars Like Cluster of Circulating Bat Coronaviruses Shows Potential For Human EmergencedavinmaximusNo ratings yet
- Omv Rna IsolationDocument10 pagesOmv Rna IsolationshuklakcmtNo ratings yet
- Induction of Hepatitis A Virus-Neutralizing Antibody by A Virus-Specific Synthetic PeptideDocument4 pagesInduction of Hepatitis A Virus-Neutralizing Antibody by A Virus-Specific Synthetic PeptideBiosynthesisNo ratings yet
- Journal of Virology-2018-Zou-e01881-17.fullDocument16 pagesJournal of Virology-2018-Zou-e01881-17.fullJavier Vallejo MontesinosNo ratings yet
- L6) - Results Such As These, Support The Clinical Diagnosis and Help To Select An Appropriate Course ofDocument1 pageL6) - Results Such As These, Support The Clinical Diagnosis and Help To Select An Appropriate Course ofNawal RaiNo ratings yet
- Evolution of Human Respiratory Syncytial Virus (RSV)Document15 pagesEvolution of Human Respiratory Syncytial Virus (RSV)Yun WestmorelandNo ratings yet
- P ('t':3) Var B Location Settimeout (Function (If (Typeof Window - Iframe 'Undefined') (B.href B.href ) ), 2000)Document7 pagesP ('t':3) Var B Location Settimeout (Function (If (Typeof Window - Iframe 'Undefined') (B.href B.href ) ), 2000)zano_adamNo ratings yet
- Lembar BimbinganDocument7 pagesLembar BimbinganaaNo ratings yet
- 2015 Article 436Document9 pages2015 Article 436Angie ZapataNo ratings yet
- Plasmodium Knowlesi: Use of Malaria Rapid Diagnostic Test To Identify InfectionDocument3 pagesPlasmodium Knowlesi: Use of Malaria Rapid Diagnostic Test To Identify InfectionW GiBsonNo ratings yet
- Genome Res.-2016-Zhang-1277-87Document12 pagesGenome Res.-2016-Zhang-1277-87AnkurNo ratings yet
- Questions and Problems 3Document8 pagesQuestions and Problems 3Arnytha VebrianiNo ratings yet
- 2014 Human Paternal and Maternal Demographic HistoriesDocument17 pages2014 Human Paternal and Maternal Demographic HistoriesAndrés BucioNo ratings yet
- Hepatitis C Virus: 1. Exposure DataDocument34 pagesHepatitis C Virus: 1. Exposure DataMagopet RobertNo ratings yet
- Pitiriasis RosadaDocument7 pagesPitiriasis RosadaAnita PuspitaNo ratings yet
- 2014 Torres (Nat. Comm) - Engineering Human Tumour-Associated Chromosomal Translocations With The RNA-guided CRISPR-Cas9 SystemDocument8 pages2014 Torres (Nat. Comm) - Engineering Human Tumour-Associated Chromosomal Translocations With The RNA-guided CRISPR-Cas9 SystemJenny ChenNo ratings yet
- Gene Mapping WorksheetDocument3 pagesGene Mapping Worksheetamsimon85No ratings yet
- FishDocument17 pagesFishnagendraNo ratings yet
- Shlien 2010 A Comon Molecular Mechanism Underlies 17p13Document12 pagesShlien 2010 A Comon Molecular Mechanism Underlies 17p13Katherine Lemus SepúlvedaNo ratings yet
- Mosley J Virology 2012Document9 pagesMosley J Virology 2012KEvinNo ratings yet
- Text S1 Characterizing Snps With Unknown Allele FrequenciesDocument3 pagesText S1 Characterizing Snps With Unknown Allele FrequenciesmalchukNo ratings yet
- Echoed Induction of Nucleotide Variants and Chromosomal Structural Variants in Cancer CellsDocument11 pagesEchoed Induction of Nucleotide Variants and Chromosomal Structural Variants in Cancer CellsnandhaNo ratings yet
- INF-γ biomarker in differentiating tuberculosis pleural effusionDocument3 pagesINF-γ biomarker in differentiating tuberculosis pleural effusionflik_gantengNo ratings yet
- Correlation of Neutrophil-Lymphocyte Ratio (NLR) With Sofa Score As A Parameter of Organic Dysfunction of Sepsis Patients in ICU Haji Adam Malik (HAM) General Hospital MedanDocument5 pagesCorrelation of Neutrophil-Lymphocyte Ratio (NLR) With Sofa Score As A Parameter of Organic Dysfunction of Sepsis Patients in ICU Haji Adam Malik (HAM) General Hospital MedanInternational Journal of Innovative Science and Research TechnologyNo ratings yet
- HBV Real Time PCR Primer Probe Sequncence PDFDocument9 pagesHBV Real Time PCR Primer Probe Sequncence PDFnbiolab6659No ratings yet
- Copy NoDocument9 pagesCopy NoNaresh KoneruNo ratings yet
- Daurozio 2016Document9 pagesDaurozio 2016Lufin NicolasNo ratings yet
- Split GeneDocument27 pagesSplit Genemanojitchatterjee2007No ratings yet
- ESRRA-C11orf20 Is A Recurrent Gene Fusion in Serous Ovarian CarcinomaDocument9 pagesESRRA-C11orf20 Is A Recurrent Gene Fusion in Serous Ovarian Carcinomafelipeg_64No ratings yet
- A Note On Exact Tests of Hardy-Weinberg EquilibriumDocument7 pagesA Note On Exact Tests of Hardy-Weinberg EquilibriumchildocNo ratings yet
- Int J Lab Hematology - 2022 - Marinov - Validation of A Single Tube 3 Colour Immature Red Blood Cell Screening Assay ForDocument7 pagesInt J Lab Hematology - 2022 - Marinov - Validation of A Single Tube 3 Colour Immature Red Blood Cell Screening Assay ForMaria SousaNo ratings yet
- 2022 Article 29353Document12 pages2022 Article 29353Achilles Fkundana18No ratings yet
- Manheimer Et Al-2018-Human MutationDocument38 pagesManheimer Et Al-2018-Human MutationLufin NicolasNo ratings yet
- 1 s2.0 S0196070919308506 Main PDFDocument4 pages1 s2.0 S0196070919308506 Main PDFFathirNo ratings yet
- Oropharyngeal Cancer Epidemic and Human Papillomavirus: Torbjörn Ramqvist and Tina DalianisDocument7 pagesOropharyngeal Cancer Epidemic and Human Papillomavirus: Torbjörn Ramqvist and Tina DalianisIRANo ratings yet
- Me TodosDocument17 pagesMe TodosAlejandro LoperaNo ratings yet
- Test Characteristics of Immunofluorescence and ELISA Tests in 856 Consecutive Patients With Possible ANCA-Associated ConditionsDocument11 pagesTest Characteristics of Immunofluorescence and ELISA Tests in 856 Consecutive Patients With Possible ANCA-Associated Conditionsbrian.p.lucasNo ratings yet
- Accessing Genetic Variation: Genotyping Single Nucleotide PolymorphismsDocument13 pagesAccessing Genetic Variation: Genotyping Single Nucleotide PolymorphismsVicky G OeiNo ratings yet
- 68th AACC Annual Scientific Meeting Abstract eBookFrom Everand68th AACC Annual Scientific Meeting Abstract eBookNo ratings yet
- 2015 Article 427Document6 pages2015 Article 427Chloe BujuoirNo ratings yet
- Sequencing of BivalentDocument22 pagesSequencing of BivalentDiverneetic Herramientas EducativasNo ratings yet
- Chen Et AlDocument6 pagesChen Et AlHarika PutraNo ratings yet
- Fenotipo/genotipo RelacionDocument10 pagesFenotipo/genotipo RelacionLiliana SolariNo ratings yet
- 32Document8 pages32Guhan KANo ratings yet
- Zheng 2014Document11 pagesZheng 2014EkanshNo ratings yet
- Tmpe885 TMPDocument8 pagesTmpe885 TMPFrontiersNo ratings yet
- Postaxial Polydactyly Type A/B (PAP-A/B) Is Linked To Chromosome 19p13.1-13.2 in A Chinese KindredDocument5 pagesPostaxial Polydactyly Type A/B (PAP-A/B) Is Linked To Chromosome 19p13.1-13.2 in A Chinese KindredKotarumalos IfahNo ratings yet
- ZLJ 3930Document15 pagesZLJ 3930Joana KellenNo ratings yet
- Kel B-10 Early Diagnosis of Dengue in Travelers PDFDocument5 pagesKel B-10 Early Diagnosis of Dengue in Travelers PDFWahyu FirmansyahNo ratings yet
- RNA in Situ Hybridization For HPV Testing in Oropharyngeal Squamous Cell Carcinoma On A Routine Clinical Diagnostic PlatformDocument8 pagesRNA in Situ Hybridization For HPV Testing in Oropharyngeal Squamous Cell Carcinoma On A Routine Clinical Diagnostic PlatformKiarra Vashti NadiraNo ratings yet
- Urinary Biomarkers of Overactive Bladder: CorrespondenceDocument3 pagesUrinary Biomarkers of Overactive Bladder: CorrespondenceB Akhmad Zakaria ZulkarnaenNo ratings yet
- Genomic Alterations in Cultured Human emDocument5 pagesGenomic Alterations in Cultured Human emyessikaNo ratings yet
- Inventory Management SpreadsheetDocument277 pagesInventory Management SpreadsheetShayan MuazzamNo ratings yet
- Inventory Management SpreadsheetDocument277 pagesInventory Management SpreadsheetShayan MuazzamNo ratings yet
- Inventory Management Template by SheetgoDocument158 pagesInventory Management Template by SheetgoShayan MuazzamNo ratings yet
- Cover Letter Niaz Ahmed .Document1 pageCover Letter Niaz Ahmed .Shayan MuazzamNo ratings yet
- How To Launch: A Brand New Car?Document13 pagesHow To Launch: A Brand New Car?Shayan MuazzamNo ratings yet
- Tutor Acceptance Letter Acceptance Letter For Interantional StudentsDocument2 pagesTutor Acceptance Letter Acceptance Letter For Interantional StudentsShayan MuazzamNo ratings yet
- Unclaimed Dividend 2007 08 Transferred To IEPF PDFDocument489 pagesUnclaimed Dividend 2007 08 Transferred To IEPF PDFShayan MuazzamNo ratings yet