Professional Documents
Culture Documents
GVsharedLOVD. Batchupload
Uploaded by
MiaOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
GVsharedLOVD. Batchupload
Uploaded by
MiaCopyright:
Available Formats
Batch upload data
Ivo Fokkema / Johan den Dunnen
For large data sets the “Global Variome shared LOVD” offers a batch upload service.
The file actually used for uploads is quite complicated, when interested you can obtain it
by using the option “Download all my data” (from the “Your account” page (after login,
top right)). We made a simplified version of this file (GVsharedLOVD_upload.xlsx).
Data in LOVD3
In LOVD3 powered databases data are collected and stored separately for the Individual,
the Phenotype, the Screening and the Variant (based on a genomic position and
optionally linked to one or more Transcripts). LOVD3 allows for one Individual to link
several different Phenotype and/or Screening entries. In addition one Screening can have
many different Variants. As a consequence, data can not be added in a file containing all
data on one line/row. We generated a simplified upload file to allow data entry where
possible per line/row. The file can be completed and mailed to
LOVD@JohanDenDunnen.nl where the LOVD-team will perform a last reformat and
take care of the actual batch upload.
Simplified upload file
The simplified upload file provided (GVsharedLOVD_upload.xlsx) should be opened in
a spread sheet program. The file contains an example data set (which should be removed)
and an explanation of specific formats used in the “Global Variome shared LOVD”.
Please make sure you complete all mandatory fields (marked in red) and use only options
from the selection list provided in the red and yellow marked fields.
NOTES
1. LOVD allows reporting data per Family as well as per Individual (our preference
is per Individual). Since most submissions contain data on isolated cases or
summary data per family, we explain such submissions only. When you want to
report several cases per family, please contact us for further details.
2. to find the {{diseaseid}} go to https://databases.lovd.nl/shared/diseases and find
the ID (general) of the Disease of the Individual reported. Select the Disease as
originally diagnosed, not of the ultimate diagnosis after a DNA analysis. For
complex cases use “00198” (unclassified / mixed). For controls use “00000”
(Healthy/Control). In {{Phenotype/Diagnosis/Initial}} give the initial general
diagnosis in words. In {{Phenotype/Diagnosis/Definite}} give the final diagnosis
as confirmed by DNA analysis (select abbreviation from https://databases.lovd.nl/
shared/diseases).
3. when more than one variant has been identified in one Individual you do not need
to repeat all information on Individual, Phenotype and Screening but only
complete details on the Variant on additional rows. When the additional Variants
were detected using another screening method, please make sure to add an
additional entry for the Screening.
You might also like
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5795)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1090)
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (838)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (895)
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (588)
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (345)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (121)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (400)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2259)
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (266)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (74)
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- Daniel Pimley - Representations of The Body in AlienDocument37 pagesDaniel Pimley - Representations of The Body in AlienVerbal_KintNo ratings yet
- 5.tang Et Al 2016Document16 pages5.tang Et Al 2016MiaNo ratings yet
- 6.fei Et Al 2015Document6 pages6.fei Et Al 2015MiaNo ratings yet
- 8.drenser Et Al 2009Document6 pages8.drenser Et Al 2009MiaNo ratings yet
- Novel Frizzled-4 Gene Mutations in Chinese Patients With Familial Exudative VitreoretinopathyDocument9 pagesNovel Frizzled-4 Gene Mutations in Chinese Patients With Familial Exudative VitreoretinopathyMiaNo ratings yet
- Archivos DE LA Sociedad Española DE OftalmologíaDocument4 pagesArchivos DE LA Sociedad Española DE OftalmologíaMiaNo ratings yet
- 2.yang Et Al 2018Document11 pages2.yang Et Al 2018MiaNo ratings yet
- 4.khan Et Al 2016Document4 pages4.khan Et Al 2016MiaNo ratings yet
- 7.dixon Et Al 2016Document4 pages7.dixon Et Al 2016MiaNo ratings yet
- Diagnostic Exome Sequencing in Persons With Severe Intellectual DisabilityDocument9 pagesDiagnostic Exome Sequencing in Persons With Severe Intellectual DisabilityMiaNo ratings yet
- 8.kuechler Et Al 2015Document13 pages8.kuechler Et Al 2015MiaNo ratings yet
- Sun Et Al 2019Document5 pagesSun Et Al 2019MiaNo ratings yet
- 11.ORoak Et Al 2012Document77 pages11.ORoak Et Al 2012MiaNo ratings yet
- 9.tucci Et Al 2014Document15 pages9.tucci Et Al 2014MiaNo ratings yet
- 4.kharbanda Et Al 2017Document6 pages4.kharbanda Et Al 2017MiaNo ratings yet
- 8.kuechler Et Al 2015Document13 pages8.kuechler Et Al 2015MiaNo ratings yet
- Wu Et Al 2016Document11 pagesWu Et Al 2016MiaNo ratings yet
- PCR Troubleshooting and OptimizationDocument608 pagesPCR Troubleshooting and Optimizationlaytailieu20220% (1)
- 5 Cover Letter Samples For Your Scientific ManuscriptDocument11 pages5 Cover Letter Samples For Your Scientific ManuscriptAlejandra J. Troncoso100% (2)
- Origin of Life 1Document2 pagesOrigin of Life 1zxcvblesterNo ratings yet
- Heredity and EvolutionDocument19 pagesHeredity and EvolutionJames Bond 007No ratings yet
- Chemistry Project On Study of Rate of Fermentation of JuicesDocument6 pagesChemistry Project On Study of Rate of Fermentation of Juicesyash dongreNo ratings yet
- Diffusion EssayDocument2 pagesDiffusion Essayapi-242424506No ratings yet
- 1 Mobilising Ca2 to enhance fruit quality Preharvest application of Harpin αβ ProAct in citrus orchards in SpainDocument7 pages1 Mobilising Ca2 to enhance fruit quality Preharvest application of Harpin αβ ProAct in citrus orchards in SpainMurilo GrecoNo ratings yet
- Lab 02 - Preparation of Smears and Gram StainingDocument10 pagesLab 02 - Preparation of Smears and Gram StainingVincent ReyesNo ratings yet
- Reading-Renaissance Learning and Unlocking Your PotentialDocument6 pagesReading-Renaissance Learning and Unlocking Your PotentialHoneyLopesNo ratings yet
- Modernage Public School & College, Abbottabad Revised Daily Grand Test Schedule & Pre-Board Date Sheet For Class 10Document1 pageModernage Public School & College, Abbottabad Revised Daily Grand Test Schedule & Pre-Board Date Sheet For Class 10Jaadi 786No ratings yet
- Hand Activities During RobberiesDocument3 pagesHand Activities During RobberiesRuben_Monroy_ClaudioNo ratings yet
- CMT Lesson 2Document14 pagesCMT Lesson 2Pam SyNo ratings yet
- Cat's Lair - Christine FeehanDocument462 pagesCat's Lair - Christine FeehanVivi Lara33% (3)
- (English (Auto-Generated) ) Debunking Myths About Adolescence (DownSub - Com)Document4 pages(English (Auto-Generated) ) Debunking Myths About Adolescence (DownSub - Com)Janine BaringNo ratings yet
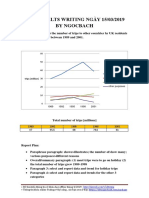
- Giai de Ielts Writing Ngay 150319 by NgocbachDocument7 pagesGiai de Ielts Writing Ngay 150319 by NgocbachSedeaNo ratings yet
- Drug Metabolism and PharmacokineticsDocument48 pagesDrug Metabolism and PharmacokineticsYunistya Dwi CahyaniNo ratings yet
- Bakir 2017Document12 pagesBakir 2017fragariavescaNo ratings yet
- Life Processes of Living Thigns (Paramecium)Document21 pagesLife Processes of Living Thigns (Paramecium)zanu winfred kwameNo ratings yet
- Compost Tea Brewing ManualDocument91 pagesCompost Tea Brewing ManualJanetS14338100% (1)
- Invigilation Time Table 2022Document3 pagesInvigilation Time Table 2022ThomasNo ratings yet
- SCI 11 Module 4Document10 pagesSCI 11 Module 4Sebastian SmytheNo ratings yet
- Brain Structures and Their FunctionsDocument6 pagesBrain Structures and Their FunctionsVasudha RohatgiNo ratings yet
- TNSCST - OldDocument15 pagesTNSCST - OldPG ChemistryNo ratings yet
- Flex Printing TenderDocument13 pagesFlex Printing TenderThe WireNo ratings yet
- 5-Environmental Chemistry and MicrobiologyDocument41 pages5-Environmental Chemistry and MicrobiologyRachelle AtienzaNo ratings yet
- Test Bank For Molecular Biology of The Cell Sixth EditionDocument30 pagesTest Bank For Molecular Biology of The Cell Sixth EditionMarie Villar100% (32)
- Reading (In-Class) 5Document6 pagesReading (In-Class) 5Trân TúNo ratings yet
- Richa Shrivastava Assistant Professor Department of Pharmacy BITS PilaniDocument18 pagesRicha Shrivastava Assistant Professor Department of Pharmacy BITS PilaniCHOUGULE KISHOR SURYAKANTNo ratings yet
- Specimen QP - Paper 1 OCR (A) Biology As-LevelDocument23 pagesSpecimen QP - Paper 1 OCR (A) Biology As-LevelNatsaisheNo ratings yet