Professional Documents
Culture Documents
Mmae 517 Ps 2 - Jupyter Notebook
Uploaded by
api-644476404Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Mmae 517 Ps 2 - Jupyter Notebook
Uploaded by
api-644476404Copyright:
Available Formats
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
Jeron Young
MMAE 517 Computational Fluid Dynamics PS #2
In [1]: import numpy as np
import scipy as sp
import scipy.linalg
import math
import matplotlib.pyplot as plt
Problem 1
In [28]: # Gauss-Seidel Iteration with SOR
N = 51
dx = 1/(N-1)
dy = 1/(N-1)
del_bar = (dx/dy)**2
x = np.linspace(0, 1, N)
y= np.linspace(0, 1, N)
m = 1
n = 1
In [29]: print(x)
print(dx)
print (del_bar)
[0. 0.02 0.04 0.06 0.08 0.1 0.12 0.14 0.16 0.18 0.2 0.22 0.24 0.26
0.28 0.3 0.32 0.34 0.36 0.38 0.4 0.42 0.44 0.46 0.48 0.5 0.52 0.54
0.56 0.58 0.6 0.62 0.64 0.66 0.68 0.7 0.72 0.74 0.76 0.78 0.8 0.82
0.84 0.86 0.88 0.9 0.92 0.94 0.96 0.98 1. ]
0.02
1.0
In [30]: # b matrix = f matrix
b = np.zeros((N, N))
for i in range(0, N):
for j in range(0, N):
b[i, j] = np.cos(np.pi*m*x[i])*np.cos(np.pi*n*y[j])
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 1/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [31]: print (b)
[[ 1. 0.99802673 0.9921147 ... -0.9921147 -0.99802673
-1. ]
[ 0.99802673 0.99605735 0.99015699 ... -0.99015699 -0.99605735
-0.99802673]
[ 0.9921147 0.99015699 0.98429158 ... -0.98429158 -0.99015699
-0.9921147 ]
...
[-0.9921147 -0.99015699 -0.98429158 ... 0.98429158 0.99015699
0.9921147 ]
[-0.99802673 -0.99605735 -0.99015699 ... 0.99015699 0.99605735
0.99802673]
[-1. -0.99802673 -0.9921147 ... 0.9921147 0.99802673
1. ]]
In [32]: b.shape
Out[32]: (51, 51)
In [33]: phi = np.zeros((N, N))
In [34]: print(phi)
[[0. 0. 0. ... 0. 0. 0.]
[0. 0. 0. ... 0. 0. 0.]
[0. 0. 0. ... 0. 0. 0.]
...
[0. 0. 0. ... 0. 0. 0.]
[0. 0. 0. ... 0. 0. 0.]
[0. 0. 0. ... 0. 0. 0.]]
In [35]: phi.shape
Out[35]: (51, 51)
In [36]: # Neumann boundary Conditons
#dphi/dx
#eqn. 5.26
c = np.zeros((N, N))
c[0, 0] = 0
c[N-1, N-1] = 0
#dphi/dy
#eqn. 5.31
d = np.zeros((N, N))
d[0, 0] = 0
d[N-1, N-1] = 0
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 2/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [37]: # 4 corners boundary conditions
#eqn. 5.30
phi[0, 0] = (1/(2*(1+del_bar)))*((2*phi[1, 0]- c[0, 0]*dx) + del_bar*(phi[0
#eqn. 5.32
phi[0, N-1] = (1/(2*(1+del_bar)))*((2*phi[1, N-1]- c[0, N-1]*dx) + 2*del_ba
#eqn. 5.32
phi[N-1, 0] = (1/(2*(1+del_bar)))*((2*phi[N-1, 1]- c[N-1, 0]*dx) + 2*del_ba
#eqn. 5.32
phi[N-1, N-1] = (1/(2*(1+del_bar)))*((2*phi[N-1, N-1] + c[N-1, N-1]*dx) + 2
In [38]: #setting up x[i], y[j] for phi matrix
#Applying neuman boundary conditions at the corners -> slide 179
# 4 sides
#single loop for i
#eqn. 5.32
for j in range(2, N-1):
phi[i, j+1] = (1/(2*(1+del_bar)))*(2*phi[2, j+1] - c[i,j]*dx) + del_bar
phi[i, j-1] = (1/(2*(1+del_bar)))*(2*phi[2, j-1] - c[i,j]*dx) + del_bar
#single loop for j
#eqn. 5.30
for i in range(2, N-3):
phi[i-1, j] = (1/2*(1+del_bar))*((2*phi[2, j+1] - c[0,j]*dx)+2*del_bar*
phi[i+1, j] = (1/2*(1+del_bar))*((2*phi[2, j] - c[0,j]*dx)+2*del_bar*(p
In [39]: epsilon = 1e-5 #residual convergence
A = np.zeros((N, N)) #Initial guess
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 3/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [ ]: epsilon =10**-4
count = 0
convergence = 1
#for q in range(0, 250):
while convergence > epsilon:
phi_old = np.copy(phi)
for i in range(2, N-1):
for j in range(2, N-1):
#neumann boundary conditions -> slide 180
#updating the boundary conditions at each iteration
#eqn 5.19
phi[i, j] = (1/2*(1+del_bar))*(phi[i+1, j]+phi[i-1, j]+ del_bar
for i in range(2, N-1):
for j in range(2, N-1):
#normalize the solution
phi[i, j] = phi[i, j] - 1/((m*np.pi)**2+(n*np.pi)**2)*(np.cos(n
count = count + 1
#convergence test -> slide 162
convergence = np.amax(abs(phi - phi_old))/np.amax(phi)
print(count)
print(convergence)
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 4/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
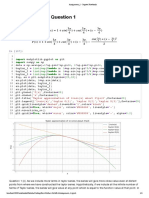
In [49]: fig = plt.figure
plt.pcolor(x, y, phi, cmap = 'jet', shading='auto')
plt.colorbar()
plt.title('Gauss-Seidel Iteration with SOR')
plt.xlabel("$x$")
plt.ylabel("$y$")
plt.show()
In [47]: #Exact Soln:
N = 51
x = np.linspace(0, 1, N)
y = np.linspace(0, 1, N)
m = 1
n = 1
X, Y = np.meshgrid(x, y)
phi = -1/((m*np.pi)**2+(n*np.pi)**2)*(np.cos(np.pi*m*X)*np.cos(np.pi*n*Y))
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 5/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [48]: fig = plt.figure
plt.pcolor(x, y, phi, cmap = 'jet', shading='auto')
plt.colorbar()
plt.title('Gauss-Seidel Iteration with SOR')
plt.xlabel("$x$")
plt.ylabel("$y$")
plt.show()
Problem 2
In [43]: #Alternating-Direction-Implict (ADI) using Thomas alg
In [45]: #51 x 51 mesh
# 51 points
N = 51
l = 1
m = 1
n = 1
x = np.linspace(0, 1, N)
y = np.linspace(0, 1, N)
dx = l/(N-1)
dy = l/(N-1)
d_bar = (dx/dy)**2
sigma = 1.1
In [46]: np.shape(x)
Out[46]: (51,)
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 6/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [73]: a = np.zeros(N)
b = np.zeros(N)
c = np.zeros(N)
d = np.zeros(N)
g = np.zeros(N)
f = np.zeros((N, N))
In [74]: np.shape(a)
Out[74]: (51,)
In [75]: # b matrix = f matrix
for i in range(0, N):
for j in range(0, N):
f[i, j] = np.cos(np.pi*m*x[i])*np.cos(np.pi*n*y[j])
In [76]: def thomas(N, dx, a, b, c, d, left, right):
F = np.zeros(N)
delta = np.zeros(N)
theta = np.zeros(N)
# Adjust coefficients for boundary conditions
b[0] = left[1]*b[0] + 2.0*dx*left[0]*a[0]
c[0] = left[1]*(c[0] + a[0])
d[0] = left[1]*d[0] + 2.0*dx*left[2]*a[0]
a[N-1] = right[1]*(a[N-1] + c[N-1])
b[N-1] = right[1]*b[N-1] - 2.0*dx*right[0]*c[N-1]
d[N-1] = right[1]*d[N-1] - 2.0*dx*right[2]*c[N-1]
# Forward elimination
F[0] = c[0]/b[0]
delta[0] = d[0]/b[0]
for k in range(1, N):
x1 = 1.0/(b[k] - a[k]*F[k-1])
F[k] = c[k]*x1
delta[k] = x1*(d[k] - a[k]*delta[k-1])
# Back substitution
theta[N-1] = delta[N-1]
for k in range(N-2,-1,-1):
theta[k] = delta[k] - F[k]*theta[k+1]
return theta
In [90]:
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 7/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [91]: print(a)
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 2.]
In [92]: count = 0
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 8/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [99]: #ADI (Neumann Boundary)
#convergence
# is phi converged?
epsilon =10**-4
count = 0
convergence = 1
while convergence > epsilon:
phi_old = np.copy(phi)
for j in range(0, N):
for i in range (0, N):
#from slide 193 -> I line iterations
#eqn. 5.36
a[i] = 1
b[i] = -(2 + sigma)
c[i] = 1
# if j = 0 then d[0]
#slide 181 (points outside of the domain) -> u[1,j] = u[2,j]
if j == 0:
for i in range (0, N):
d[0] = dx**2*f[i, j] - d_bar*(phi[i, j+1] - (2 - sigma/d_ba
elif j == N-1:
for i in range (0, N):
d[N-1] = dx**2*f[i, j] - d_bar*(phi[i, j-1] - (2 - sigma/d_
else:
for i in range (0, N):
#from slide 189 -> J line iterations (Y-lines)
#eqn. 5.35
d[i] = dx**2*f[i, j] - d_bar*(phi[i, j+1] - (2 - sigma/d_ba
# Neumann boundary conditions
# p*u + q*du/dr = r
# p=0; q=1; r = 0
left = np.array([0, 1, 0])
right = np.array([0, 1, 0])
theta0 = thomas(N, dx, a, b, c, d, left, right)
phi[:,j] = theta0
for i in range(0, N):
for j in range (0, N):
#from slide 193 -> I line iterations
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 9/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
#eqn. 5.36
a[j] = d_bar
b[j] = -(2*d_bar + sigma)
c[j] = d_bar
if i == 0:
for j in range (0, N):
d[0] = dx**2*f[i, j] - (phi[i+1, j] - (2-sigma)*phi[i, j] +
elif i == N-1:
for j in range (0, N):
d[N-1] = dx**2*f[i, j] - (phi[i-1, j] - (2-sigma)*phi[i, j]
else:
for j in range (0, N):
#from slide 193 -> I line iterations (X-lines)
#eqn. 5.36
d[j] = dx**2*f[i, j] - (phi[i+1, j] - (2-sigma)*phi[i, j] +
# Neumann boundary conditions
# q*du/dr = r
# p=0; q=1; r = 0
left = np.array([0, 1, 0])
right = np.array([0, 1, 0])
theta1 = thomas(N, dy, a, b, c, d, left, right)
phi[i,:] = theta1
count = count + 1
#convergence test -> slide 162
convergence = np.amax(abs(phi - phi_old))/np.amax(phi)
print(count)
print(convergence)
1
-0.004054612543562862
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 10/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [100]: print(phi)
[[-0.42144823 -0.42159543 -0.42053704 ... -0.43603266 -0.43639182
-0.43669493]
[-0.42056473 -0.42044099 -0.41964263 ... -0.43309712 -0.43338944
-0.43353448]
[-0.41908165 -0.4189811 -0.41842965 ... -0.42982261 -0.43006661
-0.4301613 ]
...
[-0.42336751 -0.42283381 -0.42153032 ... -0.43652483 -0.43679728
-0.43690064]
[-0.42336738 -0.4228041 -0.42148587 ... -0.43679736 -0.43709703
-0.43723749]
[-0.42336738 -0.4228041 -0.42148587 ... -0.43683166 -0.43719082
-0.43749393]]
In [101]: plt.pcolor(x, y, phi, cmap = 'jet', shading='auto')
plt.colorbar()
plt.title('ADI with Neumann Boundaries')
plt.xlabel("$x$")
plt.ylabel("$y$")
plt.show()
In [48]: #Exact Soln: (same as problem 1)
N = 51
x = np.linspace(0, 1, N)
y = np.linspace(0, 1, N)
m = 1
n = 1
X, Y = np.meshgrid(x, y)
PHI = -1/((m*np.pi)**2+(n*np.pi)**2)*(np.cos(np.pi*m*X)*np.cos(np.pi*n*Y))
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 11/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [49]: fig = plt.figure
plt.pcolor(x, y, PHI, cmap = 'jet', shading='auto')
plt.colorbar()
plt.title('ADI with Neumann Boundaries')
plt.xlabel("$x$")
plt.ylabel("$y$")
plt.show()
Problem 3
In [15]: #ADI w/ dirichelt boundary conditions
In [16]: N = 51
l = 1
x = np.linspace(0, 1, N)
y = np.linspace(0, 1, N)
dx = l/(N-1)
dy = l/(N-1)
d_bar = (dx/dy)**2
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 12/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [17]: # Iterations
I = 50
J = 50
#Acceleration Parameter -> slide 194
R = max(I +1, J + 1)
sigma = 2*np.cos(np.pi/R)
In [18]: print(sigma)
1.9962066574740882
In [19]: #Dirichelt boundary conditions
# what I did was I implemented the boundary conditions from problem 3; psi
# psi = x @ inlet and psi = y @outlet
psi = np.zeros((N, N))
for i in range(0, N):
for j in range(0, N):
psi[0, j] = 0
psi[i, 0] = 0
psi[i, N-1] = y[i]
psi[N-1, j] = x[j]
In [20]: print (psi)
[[0. 0. 0. ... 0. 0. 0. ]
[0. 0. 0. ... 0. 0. 0.02]
[0. 0. 0. ... 0. 0. 0.04]
...
[0. 0. 0. ... 0. 0. 0.96]
[0. 0. 0. ... 0. 0. 0.98]
[0. 0.02 0.04 ... 0.96 0.98 1. ]]
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 13/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [21]: def thomas(N, dx, a, b, c, d, left, right):
F = np.zeros(N)
delta = np.zeros(N)
theta = np.zeros(N)
# Adjust coefficients for boundary conditions
b[0] = left[1]*b[0] + 2.0*dx*left[0]*a[0]
c[0] = left[1]*(c[0] + a[0])
d[0] = left[1]*d[0] + 2.0*dx*left[2]*a[0]
a[N-1] = right[1]*(a[N-1] + c[N-1])
b[N-1] = right[1]*b[N-1] - 2.0*dx*right[0]*c[N-1]
d[N-1] = right[1]*d[N-1] - 2.0*dx*right[2]*c[N-1]
# Forward elimination
F[0] = c[0]/b[0]
delta[0] = d[0]/b[0]
for k in range(1, N):
x1 = 1.0/(b[k] - a[k]*F[k-1])
F[k] = c[k]*x1
delta[k] = x1*(d[k] - a[k]*delta[k-1])
# Back substitution
theta[N-1] = delta[N-1]
for k in range(N-2,-1,-1):
theta[k] = delta[k] - F[k]*theta[k+1]
return theta
In [22]: a = np.zeros(N)
b = np.zeros(N)
c = np.zeros(N)
d = np.zeros(N)
g = np.zeros(N)
f = np.zeros((N, N))
In [23]: for j in range(0, N):
#from slide 193 -> y line iterations
#eqn. 5.36
a[j] = 1
b[j] = -(2 + sigma)
c[j] = 1
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 14/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [24]: for i in range(0, N):
#from slide 193 -> x line iterations
#eqn. 5.36
a[i] = d_bar
b[i] = -(2*d_bar + sigma)
c[i] = d_bar
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 15/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [25]: #ADI
epsilon =10**-4
count = 0
convergence = 1
#for q in range(0, 250):
while convergence > epsilon:
psi_old = np.copy(psi)
for j in range(1, N-1):
#from slide 189 -> J line iterations
#eqn. 5.35
for i in range (1, N-1):
d[i] = dx**2*f[i, j] - d_bar*(psi[i, j+1] - (2 - sigma/d_bar)*p
# Dirichlet boundary conditions
# q*du/dr = r
# p=0; q=1; r = rhs (right hand side)
left = np.array([1, 0, 0])
right = np.array([1, 0, y[j]])
theta0 = thomas(N, dx, a, b, c, d, left, right)
psi[:,j] = theta0
for i in range(1, N-1):
#from slide 193 -> I line iterations
#eqn. 5.36
for j in range (1, N-1):
d[j] = dx**2*f[i, j] - (psi[i+1, j] - (2-sigma)* psi[i, j] + ps
# Dirichelt boundary conditions
# q*du/dr = r
# p=1; q=0; r = rhs (right hand side)
left = np.array([1, 0, 0])
right = np.array([1, 0, x[i]])
theta1 = thomas(N, dx, a, b, c, d, left, right)
psi[i,:] = theta1
count = count + 1
#convergence test -> slide 162
convergence = np.amax(abs(psi - psi_old))/np.amax(psi)
print(count)
print(convergence)
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 16/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
268
9.904490812648326e-05
In [94]: plt.pcolor(x, y, psi, cmap = 'jet', shading='auto')
plt.colorbar()
plt.title('ADI w/ Dirichlet boundaries')
plt.xlabel("$x$")
plt.ylabel("$y$")
plt.show()
In [10]: #Exact Soln
N = 51
x = np.linspace(0, 1, N)
y = np.linspace(0, 1, N)
In [11]: X, Y = np.meshgrid(x, y)
PSI = X*Y
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 17/18
12/7/22, 9:37 AM MMAE 517 PS #2 - Jupyter Notebook
In [12]: fig = plt.figure
plt.pcolor(X, Y, PSI, cmap = 'jet', shading='auto') # Pseudocolor
plt.colorbar() # Include colorbar on plot
plt.title('ADI w/ Dirichlet boundaries')
plt.show()
In [ ]:
localhost:8888/notebooks/MMAE 517 PS %232.ipynb 18/18
You might also like
- Analytic Geometry: Graphic Solutions Using Matlab LanguageFrom EverandAnalytic Geometry: Graphic Solutions Using Matlab LanguageNo ratings yet
- Answers to Selected Problems in Multivariable Calculus with Linear Algebra and SeriesFrom EverandAnswers to Selected Problems in Multivariable Calculus with Linear Algebra and SeriesRating: 1.5 out of 5 stars1.5/5 (2)
- Mmae 517 Ps 1 - Jupyter NotebookDocument15 pagesMmae 517 Ps 1 - Jupyter Notebookapi-644476404No ratings yet
- SorDocument3 pagesSorJimmy OmarNo ratings yet
- W5 Ps Fourier Acoustic 2dDocument7 pagesW5 Ps Fourier Acoustic 2dvitucho92No ratings yet
- Andesnia Qonita Luthfiya - Assignment3Document13 pagesAndesnia Qonita Luthfiya - Assignment3EcicieeNo ratings yet
- Linear Algebra For Quantum Computing (From Amelie Schreiber Notebook)Document72 pagesLinear Algebra For Quantum Computing (From Amelie Schreiber Notebook)Jan_SuNo ratings yet
- Señales y Sistemas - UNPRGDocument10 pagesSeñales y Sistemas - UNPRGSlo BluckNo ratings yet
- ### Usaremos A T Dentro Del Intervalo - 2 Pi, 2 PiDocument3 pages### Usaremos A T Dentro Del Intervalo - 2 Pi, 2 PiArmenta YaiirNo ratings yet
- Assignment 1 Utkarsh - Jupyter NotebookDocument4 pagesAssignment 1 Utkarsh - Jupyter NotebookDeepankar SinghNo ratings yet
- Matlab Tips and Tricks: Gabriel Peyr e Peyre@cmapx - Polytechnique.fr August 10, 2004Document10 pagesMatlab Tips and Tricks: Gabriel Peyr e Peyre@cmapx - Polytechnique.fr August 10, 2004Raymond PraveenNo ratings yet
- Assignment 3Document8 pagesAssignment 3MadhavNo ratings yet
- Python Script For Static Deflection of A Beam Using Finite Elements - Mechanics and MachinesDocument5 pagesPython Script For Static Deflection of A Beam Using Finite Elements - Mechanics and MachinesVarghese MathewNo ratings yet
- BVP FDM 2024Document3 pagesBVP FDM 2024getachewbonga09No ratings yet
- Pro1 To 4 and 6 To 7Document13 pagesPro1 To 4 and 6 To 7Prajwal P GNo ratings yet
- Assignment 1 PythonDocument1 pageAssignment 1 PythonAashish AryaNo ratings yet
- GraphDocument4 pagesGraphRakesh M BNo ratings yet
- Graph Convolutional Network (GCN) for Karate Club Node ClassificationDocument4 pagesGraph Convolutional Network (GCN) for Karate Club Node ClassificationRakesh M BNo ratings yet
- Import As: Cal - Pivot Array ArrayDocument12 pagesImport As: Cal - Pivot Array ArraySanodariya Kshitij Ashvinchandra (B19ME075)No ratings yet
- Uas Fisika KomputasiDocument3 pagesUas Fisika KomputasiBaiq Restiyalis KurniaNo ratings yet
- TP: Algorithme Dijkstra: 1. Implémentation Du GrapheDocument4 pagesTP: Algorithme Dijkstra: 1. Implémentation Du Graphethe knowledge is powerNo ratings yet
- FFTDocument6 pagesFFTIhtisham UddinNo ratings yet
- Laboratory 11 Analysis of Digital Filters: ObjectivesDocument11 pagesLaboratory 11 Analysis of Digital Filters: ObjectivesFaraz AbbasNo ratings yet
- Python PracticalDocument30 pagesPython PracticalSayan MajiNo ratings yet
- Mathlab 5Document7 pagesMathlab 5Rakshitha ManjunathNo ratings yet
- NC Lab 07Document3 pagesNC Lab 07kuripunjabi066No ratings yet
- me232a2Document2 pagesme232a2Anubhav AgrawalNo ratings yet
- US Airports: Usairport (Http://konect - Uni-Koblenz - De/networks/opsahl-Usairport)Document3 pagesUS Airports: Usairport (Http://konect - Uni-Koblenz - De/networks/opsahl-Usairport)aniketNo ratings yet
- Tugas - Metnum - Archyuda FarchanDocument8 pagesTugas - Metnum - Archyuda FarchanArchyuda FarchanNo ratings yet
- NC Post 007Document20 pagesNC Post 007Xyed Haider BukhariNo ratings yet
- Microwave Engineering Pozar 06 Exercise 12Document6 pagesMicrowave Engineering Pozar 06 Exercise 12John Bofarull GuixNo ratings yet
- CSC323 Sp2016 QB Module 1 Efficiency of AlgorithmsDocument14 pagesCSC323 Sp2016 QB Module 1 Efficiency of AlgorithmsAhmed FawzyNo ratings yet
- BECOB236 CodeDocument10 pagesBECOB236 CodekpNo ratings yet
- Problem Set 5: MAS160: Signals, Systems & Information For Media TechnologyDocument10 pagesProblem Set 5: MAS160: Signals, Systems & Information For Media TechnologyLulzim LumiNo ratings yet
- Regression: 1 Implementación de Métodos de RegresiónDocument11 pagesRegression: 1 Implementación de Métodos de RegresiónÓscar Alfonso Gómez SepúlvedaNo ratings yet
- HW 1Document29 pagesHW 1Oscar VidesNo ratings yet
- MATLAB Inverse Z-TransformDocument6 pagesMATLAB Inverse Z-TransformAmal NairNo ratings yet
- AIML Lab ManualDocument9 pagesAIML Lab ManualAARUNI RAINo ratings yet
- Analisis Dinamico Eje XDocument24 pagesAnalisis Dinamico Eje XVICTOR MANUEL PAITAN MENDEZNo ratings yet
- Brute-Force Matching With ORB DescriptorsDocument7 pagesBrute-Force Matching With ORB DescriptorsAdib Ahmad IstiqlalNo ratings yet
- Q1 (2) - Jupyter NotebookDocument3 pagesQ1 (2) - Jupyter NotebookXYZNo ratings yet
- UntitledDocument2 pagesUntitledsatadru sinhaNo ratings yet
- Matrice2 PyDocument2 pagesMatrice2 PyLamiss GhoulNo ratings yet
- Jupyter Notebook Iterative MethodDocument4 pagesJupyter Notebook Iterative MethodJuan GuerreroNo ratings yet
- Python Final Lab 2019Document35 pagesPython Final Lab 2019vanithaNo ratings yet
- DWDM LAB-2 Matrix Multiplication Time ComparisonDocument4 pagesDWDM LAB-2 Matrix Multiplication Time Comparisonrupa sreeNo ratings yet
- Lec 4 Complete Amortize Loan 01102023 113433amDocument6 pagesLec 4 Complete Amortize Loan 01102023 113433ambdbhbNo ratings yet
- Class Def Float Float Float Def Return Def Import Return Def ReturnDocument3 pagesClass Def Float Float Float Def Return Def Import Return Def ReturnHoucemeddine FilaliNo ratings yet
- CS Lab ProgramsDocument52 pagesCS Lab Programsyerrakulathanmayi2004No ratings yet
- test_solver (3)Document2 pagestest_solver (3)mmvkebgucpwxrouzqlNo ratings yet
- 8 Animation Parabole - PyDocument3 pages8 Animation Parabole - PyDoan HunterNo ratings yet
- Import MaDocument4 pagesImport Mahulinyong1210No ratings yet
- CorrDocument6 pagesCorrchrisNo ratings yet
- Prac 4)Document1 pagePrac 4)nameistushar12No ratings yet
- Exp 4-7Document26 pagesExp 4-7ShahanasNo ratings yet
- Build Deep Neural Networks Step-by-StepDocument44 pagesBuild Deep Neural Networks Step-by-StepjiaNo ratings yet
- P Nao e Menor Que 2Document4 pagesP Nao e Menor Que 2Cíntia PimentelNo ratings yet
- Digital Signal Processing Lab task outputs and answersDocument18 pagesDigital Signal Processing Lab task outputs and answersMuhammad AnasNo ratings yet
- September 30, 2020: This Exercise Covers The Following AspectsDocument9 pagesSeptember 30, 2020: This Exercise Covers The Following AspectsSarah MendesNo ratings yet
- Inverse Trigonometric Functions (Trigonometry) Mathematics Question BankFrom EverandInverse Trigonometric Functions (Trigonometry) Mathematics Question BankNo ratings yet
- Codr Aiaa CompliantDocument39 pagesCodr Aiaa Compliantapi-644476404No ratings yet
- Design FinalDocument15 pagesDesign Finalapi-644476404No ratings yet
- Mmae 472 Research PaperDocument11 pagesMmae 472 Research Paperapi-644476404No ratings yet
- 2018dbf Illinois Institute of Technology Design ReportDocument42 pages2018dbf Illinois Institute of Technology Design Reportapi-644476404No ratings yet
- Short Ipro Presentation Spring 2021Document18 pagesShort Ipro Presentation Spring 2021api-644476404No ratings yet
- Fact SheetDocument1 pageFact Sheetapi-644476404No ratings yet
- Preliminary Design PresentationDocument26 pagesPreliminary Design Presentationapi-644476404No ratings yet
- Project Charter Budget WorksheetDocument6 pagesProject Charter Budget WorksheetnsadnanNo ratings yet
- Python MultiprocessingDocument14 pagesPython MultiprocessingUddayan SharmaNo ratings yet
- CSC 101 Assignment 1Document13 pagesCSC 101 Assignment 1Adil MalikNo ratings yet
- CONSISTENT CHECKPOINTSDocument26 pagesCONSISTENT CHECKPOINTSSwati MishraNo ratings yet
- i.MX 6 Series USB Certification GuideDocument126 pagesi.MX 6 Series USB Certification GuideXaoc KabataNo ratings yet
- Client Side Scripting (CSS) (22519) Semester - V (CO) : A Laboratory Manual ForDocument45 pagesClient Side Scripting (CSS) (22519) Semester - V (CO) : A Laboratory Manual ForIsha RangariNo ratings yet
- Dimetra Xcore Data SheetDocument12 pagesDimetra Xcore Data SheetmosaababbasNo ratings yet
- Trend Next DumpsDocument129 pagesTrend Next DumpspraveenaNo ratings yet
- Getting Started MIDASDocument271 pagesGetting Started MIDASpradeepjoshi007No ratings yet
- SQL Advanced SQL Query Optimization Techniques B09978MXWXDocument125 pagesSQL Advanced SQL Query Optimization Techniques B09978MXWXmoon26100% (1)
- TNM Car PDFDocument19 pagesTNM Car PDFSorin IonesiNo ratings yet
- Dashboards for Automobile Data VisualizationDocument2 pagesDashboards for Automobile Data VisualizationekeneNo ratings yet
- Instructor: Craig Duckett: Stored Procedures (SQL Server) MysqlDocument72 pagesInstructor: Craig Duckett: Stored Procedures (SQL Server) MysqlPhong Thien ThanhNo ratings yet
- Program Splash Screen Starts Up and Then Closes (Stand-Alone) - AutoCAD - Autodesk Knowledge NetworkDocument4 pagesProgram Splash Screen Starts Up and Then Closes (Stand-Alone) - AutoCAD - Autodesk Knowledge NetworkFaizul ZainudinNo ratings yet
- Attribution in Cyberspace Techniques andDocument15 pagesAttribution in Cyberspace Techniques andPanna AngelikaNo ratings yet
- Change Control Request Form Change Request Number:: General InformationDocument2 pagesChange Control Request Form Change Request Number:: General Informationarunrpatil0465450% (1)
- Introduction To Corel DrawDocument57 pagesIntroduction To Corel DrawOkolo Ebuka Gentility100% (2)
- Fire Alarm BookDocument196 pagesFire Alarm Bookprasanth_kb100% (4)
- Beyond The Hype: A Guide To Understanding and Successfully Implementing Artificial Intelligence Within Your BusinessDocument20 pagesBeyond The Hype: A Guide To Understanding and Successfully Implementing Artificial Intelligence Within Your BusinessNitin RamnaniNo ratings yet
- Uas Pemograman MobileDocument10 pagesUas Pemograman MobileL GoatNo ratings yet
- AEIOU CanvasDocument1 pageAEIOU CanvasKripalsinh ChudasamaNo ratings yet
- TOEFL iBT-The Test AdministrationDocument10 pagesTOEFL iBT-The Test AdministrationIemo LuHerreraNo ratings yet
- P543&5 en PM A22Document48 pagesP543&5 en PM A22GuirosilvaNo ratings yet
- Consolidated PSV Catalog PDFDocument481 pagesConsolidated PSV Catalog PDFaxiomataNo ratings yet
- SE Quote on Life DirectionDocument1 pageSE Quote on Life Directiontrols sadNo ratings yet
- Ebook GuideToOpenTelemetryDocument17 pagesEbook GuideToOpenTelemetryDavid UzielNo ratings yet
- Data Science Course Syllabus 01Document20 pagesData Science Course Syllabus 01Ram Mohan Reddy Putta100% (1)
- The Operating System's JobDocument30 pagesThe Operating System's JobGlyndel D DupioNo ratings yet
- Solution To Multi-Depot Vehicle Routing Problem Using Genetic AlgorithmsDocument14 pagesSolution To Multi-Depot Vehicle Routing Problem Using Genetic AlgorithmsTI Journals PublishingNo ratings yet
- CountingDocument2 pagesCountingDave Jedidiah TibaldoNo ratings yet