Professional Documents
Culture Documents
Lec 3 Mam Sadia
Uploaded by
hidayatullahbjr990 ratings0% found this document useful (0 votes)
11 views5 pagesTranscription in prokaryotes involves several key sequences and processes. The Pribnow box, located 10 bases before the transcription start site, is where DNA melting occurs to form the transcription bubble. The -35 sequence interacts with RNA polymerase 35 bases before transcription starts. Termination involves a Rho factor that destabilizes the RNA-DNA hybrid. Some antibiotics like rifampin and actinomycin D can inhibit transcription. Eukaryotic RNAs undergo processing including 5' capping, 3' polyadenylation, and splicing of introns from mRNA.
Original Description:
Original Title
lec 3 mam sadia
Copyright
© © All Rights Reserved
Available Formats
PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentTranscription in prokaryotes involves several key sequences and processes. The Pribnow box, located 10 bases before the transcription start site, is where DNA melting occurs to form the transcription bubble. The -35 sequence interacts with RNA polymerase 35 bases before transcription starts. Termination involves a Rho factor that destabilizes the RNA-DNA hybrid. Some antibiotics like rifampin and actinomycin D can inhibit transcription. Eukaryotic RNAs undergo processing including 5' capping, 3' polyadenylation, and splicing of introns from mRNA.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
11 views5 pagesLec 3 Mam Sadia
Uploaded by
hidayatullahbjr99Transcription in prokaryotes involves several key sequences and processes. The Pribnow box, located 10 bases before the transcription start site, is where DNA melting occurs to form the transcription bubble. The -35 sequence interacts with RNA polymerase 35 bases before transcription starts. Termination involves a Rho factor that destabilizes the RNA-DNA hybrid. Some antibiotics like rifampin and actinomycin D can inhibit transcription. Eukaryotic RNAs undergo processing including 5' capping, 3' polyadenylation, and splicing of introns from mRNA.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
You are on page 1of 5
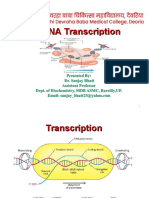
Transcription (contd)
• DNA template strand = Antisense , negative sense
• DNA coding strand = Sense , positive sense
• Consensus Sequence = A consensus sequence is a sequence
of DNA, RNA, or protein that represents aligned, related
sequences. These sequences are recognized by sigma factor .
Egs are pribnow box and -35 sequence
• The Pribnow box is a sequence of TATAAT of six nucleotides
that is an essential part of a promoter site on DNA for
transcription to occur in bacteria.
Dr.Sadia Ghousia Baig 1
Pribnow box
• Located at -10 from the transcription start site (+1).
• Site of melting (unwinding) of a short stretch ( ¬14 bp) of DNA
• Melting forms transcription bubble
-35 Sequence
• Consensus sequence 5′-TTGACA-3′
• 35 bases to the left of transcription start site
• Initial point of contact for holoenzyme
• A base is assigned negative number if it occurs prior to
transcription start site
Dr.Sadia Ghousia Baig 2
Rho Factor(p)
• The rho factor is a protein which recognizes and binds to termination sequences
in the 3′ portion of the RNA. This binding destabilizes the polymerase-template
interaction leading to dissociation of the polymerase and termination of
transcription.
• It is a hexameric ATPase with helicase activity. Separates RNA-DNA hybrid helix
causing release of RNA
• P independent termination
• Some sequences of nascent RNA are self complementary & RNA folds back on
itself forming GC rich stem and a loop ( hairpin ). Beyond hair pin RNA transcript
contains string of U’s at 3′ end. The formation of the stem-loop in the RNA
destabilizes the association between polymerase and the DNA template. This is
further destabilized by the weaker nature of the AU base pairs that are formed,
between the template and the RNA, following the stem-loop. This leads to
dissociation of polymerase and termination of transcription.
Dr.Sadia Ghousia Baig 3
Antibiotics which inhibit RNA synthesis
• Rifampin ( treats tuberculosis)
• Binds with beta subunit of prokaryotic RNA pol and prevent
chain growth beyond 3 nucleotides
• Dactinomycin (actinomycin D)
• Treats tumors
• Inserts (intercalate) between DNA bases inhibit transcription
• Initiation and elongation both inhibit
Dr.Sadia Ghousia Baig 4
Post-transcriptional Processing of RNAs
• Eukaryotic RNAs (all three classes) undergo significant processing, some of which occurs co-
transcriptionally and some post-transcriptionally
1) mRNA 5′-End Capping
• The capping process occurs after the newly synthesizing mRNA is around 20–30 bases long, at
which point RNA pol II pauses on the template.The 5′ ends of nearly all eukaryotic mRNAs are
capped with a unique 5′ → 5′ triphosphate linkage to a 7-methylguanosine residue. Synthesis of
the mRNA cap structure is catalyzed by RNA guanylyltransferase and 5′-phosphatase
• The capped end of the mRNA is protected from exonucleases & is recognized by specific
proteins of the translational machinery.
2) mRNA 3′-End Polyadenylation
• Almost all mammalian mRNAs are polyadenylated at the 3′-end.
• A specific sequence, AAUAAA, is the primary poyadenylation signal sequence . Polyadenylate
polymerase is the enzyme involved. 40-250 adenosine monophosphates are attached .Tailing
helps to stabilize mRNA and aid in translation.
3) Splicing
• Maturation of eukaryotic mRNA requires removal of RNA sequences that do not code for proteins (
introns or intervening sequences)
• The remaining coding sequences ( exons ) are joined together
• Process of removing introns and joining exons is called splicing
Dr.Sadia Ghousia Baig 5
You might also like
- Student Exploration Building DNADocument5 pagesStudent Exploration Building DNASidemen For Life32% (38)
- Microbiology An Introduction 13th Edition Tortora Test BankDocument24 pagesMicrobiology An Introduction 13th Edition Tortora Test Bankeugenephedrayy2bfk100% (39)
- PCR Amplification Lab ReportDocument5 pagesPCR Amplification Lab ReportWilson Chan100% (7)
- RNA Synthesis and ProcessingDocument17 pagesRNA Synthesis and ProcessingInnocent Clifford MaranduNo ratings yet
- TranscriptionDocument61 pagesTranscriptiondeepak mauryaNo ratings yet
- MB Chapter 5 TranscriptionDocument32 pagesMB Chapter 5 TranscriptionMustee TeferaNo ratings yet
- Transcription: DR Vaishali Dhat Professor, Department of Biochemistry DR DypmcDocument25 pagesTranscription: DR Vaishali Dhat Professor, Department of Biochemistry DR Dypmcnishkarsh chauhanNo ratings yet
- TranscriptionDocument34 pagesTranscriptiondrhydrogenNo ratings yet
- TranscriptionDocument10 pagesTranscriptionHardik ManekNo ratings yet
- RNA Processing or Post Transcriptional Modifications.Document23 pagesRNA Processing or Post Transcriptional Modifications.saeed313bbtNo ratings yet
- Transcription: Dr. Anirudra Gurung, Department of Botany, St. Joseph's College, DarjeelingDocument9 pagesTranscription: Dr. Anirudra Gurung, Department of Botany, St. Joseph's College, DarjeelingRahul JaiswalNo ratings yet
- DNA Transcription (Part-1) : DR - Ahmed Salim Mohammed PH.D Molecular MicrobiologyDocument49 pagesDNA Transcription (Part-1) : DR - Ahmed Salim Mohammed PH.D Molecular MicrobiologyDrAhmedSalimNo ratings yet
- Transcription and RegulationDocument40 pagesTranscription and RegulationRUDRANSH PUJARINo ratings yet
- Central-Dogma 2Document22 pagesCentral-Dogma 2Milena De CresentNo ratings yet
- Transcription: M.Prasad Naidu MSC Medical Biochemisty, Ph.D.Research ScholarDocument33 pagesTranscription: M.Prasad Naidu MSC Medical Biochemisty, Ph.D.Research ScholarDr. M. Prasad NaiduNo ratings yet
- 25 LN RNA Organization and Transcription CNRAADocument55 pages25 LN RNA Organization and Transcription CNRAADakshitha DharmakeerthiNo ratings yet
- Central Dogma: Faculty of Medicine, Sriwijaya UniversityDocument51 pagesCentral Dogma: Faculty of Medicine, Sriwijaya UniversityIlsya PertiwiNo ratings yet
- Gene ExpressionDocument72 pagesGene ExpressionMika ForwardNo ratings yet
- Gene Expression - Sept 2012Document36 pagesGene Expression - Sept 2012rositaNo ratings yet
- Biochemistry: RNA Synthesis and ProcessingDocument54 pagesBiochemistry: RNA Synthesis and ProcessingAqsa YaminNo ratings yet
- Post-Transcriptional Modifications: Arun Kumar PHD Scholar Centre For Molecular Biology Central University of JammuDocument67 pagesPost-Transcriptional Modifications: Arun Kumar PHD Scholar Centre For Molecular Biology Central University of JammuARUN KUMARNo ratings yet
- UntitledDocument45 pagesUntitledjemNo ratings yet
- Transcription WorkingDocument51 pagesTranscription Workingapi-3858544No ratings yet
- Gene Expression: Vikrant Joshi F.Y. Biotechnology 6519 Khalsa College AmritsarDocument31 pagesGene Expression: Vikrant Joshi F.Y. Biotechnology 6519 Khalsa College Amritsarvikrant4018No ratings yet
- Chapter 5 Protein SynthesisDocument26 pagesChapter 5 Protein SynthesisT MokshithaNo ratings yet
- 21 TranscriptionDocument39 pages21 TranscriptioneraasyahirahNo ratings yet
- Kuliah 8. Transcription (Oke)Document43 pagesKuliah 8. Transcription (Oke)Sinta DosiNo ratings yet
- 11 Chapter 5Document38 pages11 Chapter 5toobashafiNo ratings yet
- Topic 10 Transcription Lecture NotesDocument33 pagesTopic 10 Transcription Lecture NoteskambulukatambilaiNo ratings yet
- 5 11transcription-2013Document30 pages5 11transcription-2013jernsssNo ratings yet
- Lecture 2 Genomes, Cloning Part 1 AP0701 2023-24Document122 pagesLecture 2 Genomes, Cloning Part 1 AP0701 2023-24suvetha.bawaniNo ratings yet
- Nucleic Acid Metabolism TAUDocument38 pagesNucleic Acid Metabolism TAUFaatimah EssaNo ratings yet
- Transcription and Post TranscriptionDocument45 pagesTranscription and Post TranscriptionAnne Gabrielle Marapon AgsunodNo ratings yet
- Base Alterations and Base DamageDocument22 pagesBase Alterations and Base Damagemasthan6yNo ratings yet
- TranscriptionDocument23 pagesTranscriptionareen fakhouryNo ratings yet
- Gene ExpressionDocument19 pagesGene Expressionkvicto100% (1)
- 106 Ceng w10Document62 pages106 Ceng w10mertNo ratings yet
- G. Gene Expression and Protein SynthesisDocument50 pagesG. Gene Expression and Protein SynthesisMary Rose Bobis VicenteNo ratings yet
- Mrs. Ofelia Solano Saludar: Department of Natural Sciences University of St. La Salle Bacolod CityDocument55 pagesMrs. Ofelia Solano Saludar: Department of Natural Sciences University of St. La Salle Bacolod CitymskikiNo ratings yet
- Lecture 3Document30 pagesLecture 3benadethamwalusanyaNo ratings yet
- 22 (W13) TranscriptionDocument38 pages22 (W13) TranscriptionMUHAMMAD FAHMI MOHD ZAMZANINo ratings yet
- Dna TranscriptonDocument26 pagesDna TranscriptonJesse Kate GonzalesNo ratings yet
- TranscriptionDocument100 pagesTranscriptionSreshttNo ratings yet
- Dna L13 NotesDocument6 pagesDna L13 NotesellieNo ratings yet
- TranscriptionDocument64 pagesTranscriptionNasroedien FikryNo ratings yet
- Dna L12 NotesDocument6 pagesDna L12 NotesellieNo ratings yet
- LECTURE 12 TranslationDocument17 pagesLECTURE 12 TranslationAditi SharmaNo ratings yet
- Bala Sir Transcrption PDFDocument9 pagesBala Sir Transcrption PDFAnjali Ak GuptaNo ratings yet
- 3rd Week (1) - TrancriptionDocument46 pages3rd Week (1) - TrancriptionyazicigaamzeNo ratings yet
- Biochemistry Lecture PPT 11Document7 pagesBiochemistry Lecture PPT 11Chiranjeevi JoshiNo ratings yet
- Biochemistry - Biosynthesis of Protein & Protein ChemistryDocument113 pagesBiochemistry - Biosynthesis of Protein & Protein ChemistryAdella AnggrainiNo ratings yet
- Transcription 2Document66 pagesTranscription 2Safe BoxNo ratings yet
- Unit 6. Proteins SynthesisDocument57 pagesUnit 6. Proteins Synthesismunyanezaolivier422No ratings yet
- Lecture 2Document33 pagesLecture 2SUNDAS FATIMANo ratings yet
- Rna Biosynthesis (Transicription)Document33 pagesRna Biosynthesis (Transicription)Alaa AlmajedNo ratings yet
- 2 TranscriptionDocument51 pages2 Transcriptionmaksumrabbi rumc1999No ratings yet
- Transcription, Is The Process of Creating An EquivalentDocument7 pagesTranscription, Is The Process of Creating An EquivalentSandarbh GoswamiNo ratings yet
- Gene Expression 1Document28 pagesGene Expression 1ha88ial88No ratings yet
- Lecture 11 TranscriptionDocument21 pagesLecture 11 TranscriptionAditi SharmaNo ratings yet
- Transcription: From DNA To RNADocument74 pagesTranscription: From DNA To RNAmd habibur rahmanNo ratings yet
- Protein-Synthesis Online 1st ClassDocument43 pagesProtein-Synthesis Online 1st ClassAkasha khanNo ratings yet
- LECTURE 9-Transgenic Technology in Marine Organisms PDFDocument26 pagesLECTURE 9-Transgenic Technology in Marine Organisms PDFIntan Lestari DewiNo ratings yet
- PCRDocument42 pagesPCRwulanopratamiNo ratings yet
- Kami Export - RNA HIVDocument2 pagesKami Export - RNA HIVEva EllisNo ratings yet
- Tesis doctoral de Bryan Keith Holland - “Discovery of mature microRNA sequences within the protein-coding regions of global HIV-1 genomes - Predictions of novel mechanisms for viral infection and pathogenicity”Document183 pagesTesis doctoral de Bryan Keith Holland - “Discovery of mature microRNA sequences within the protein-coding regions of global HIV-1 genomes - Predictions of novel mechanisms for viral infection and pathogenicity”Omar Alejandro Herrera ArenasNo ratings yet
- Bif501 Assignment No 2Document10 pagesBif501 Assignment No 2bc190205832 RIFFAT BANONo ratings yet
- Topic 6 Gene Control: Suggested ActivitiesDocument2 pagesTopic 6 Gene Control: Suggested ActivitiesSemwezi EnockNo ratings yet
- New Developments in 454 Sequencing and The Future of Sequencing at RocheDocument31 pagesNew Developments in 454 Sequencing and The Future of Sequencing at RocheKtiapcyio DooohyxzNo ratings yet
- Keiser University Online Microbiology I Virtual Lab 4 Identification of Bacteria by Conventional & DNA TestingDocument5 pagesKeiser University Online Microbiology I Virtual Lab 4 Identification of Bacteria by Conventional & DNA TestingPrincess Lheakyrie CasilaoNo ratings yet
- Polymerase Chain Reaction (PCR) - Principle, Steps, ApplicationsDocument5 pagesPolymerase Chain Reaction (PCR) - Principle, Steps, ApplicationsMokhtarCheikhNo ratings yet
- Transcription and RegulationDocument40 pagesTranscription and RegulationRUDRANSH PUJARINo ratings yet
- Analytical Biochemistry 1990 Engelke Taq PurificationDocument5 pagesAnalytical Biochemistry 1990 Engelke Taq PurificationPablo L. Cossio RodriguezNo ratings yet
- Cooperative Learning Lesson PlanDocument2 pagesCooperative Learning Lesson Planapi-248365515No ratings yet
- Genetic Code 12Document16 pagesGenetic Code 12Jahnavi JoshiNo ratings yet
- Accurate QuantificationDocument8 pagesAccurate QuantificationANGIE TATIANA RUIZ LADINONo ratings yet
- BiologyDocument24 pagesBiologyTom KiloitNo ratings yet
- Forensic DNA Analysis Y-STRDocument10 pagesForensic DNA Analysis Y-STRWawan BwNo ratings yet
- Topic 10 Transcription Lecture NotesDocument33 pagesTopic 10 Transcription Lecture NoteskambulukatambilaiNo ratings yet
- DNA Sequencing (Article) - Biotechnology - Khan AcademyDocument8 pagesDNA Sequencing (Article) - Biotechnology - Khan AcademyZannat AraNo ratings yet
- 02-Nucleic Acids - Structure and FunctionDocument70 pages02-Nucleic Acids - Structure and FunctionBerru VardarNo ratings yet
- DNA Vs RNA - Introduction and Differences Between DNA and RNADocument10 pagesDNA Vs RNA - Introduction and Differences Between DNA and RNAKienlevyNo ratings yet
- Infographic (Lab 1 - Lab 5) AS1144A2Document1 pageInfographic (Lab 1 - Lab 5) AS1144A2Nawar SallNo ratings yet
- Protein-DNA Interaction - Wikipedia, The Free EncyclopediaDocument2 pagesProtein-DNA Interaction - Wikipedia, The Free EncyclopediaShailendra YadavNo ratings yet
- Diphenylamin e Test For Deoxyribose By: Anne Marielle D. ManaloDocument17 pagesDiphenylamin e Test For Deoxyribose By: Anne Marielle D. ManaloAcel Jone CayotNo ratings yet
- Earth and Life Science: Quarter 2 - Module 4Document20 pagesEarth and Life Science: Quarter 2 - Module 4Jerald De La CruzNo ratings yet
- Assignment GeneticDocument5 pagesAssignment GeneticAinAzmiNo ratings yet
- MY 1st Sem. Presttn DNA Damage & Repair PPT MW 2003Document26 pagesMY 1st Sem. Presttn DNA Damage & Repair PPT MW 2003Samhita Kalita100% (1)
- Chapter 8 HomeworkDocument4 pagesChapter 8 HomeworkKvn4N6No ratings yet