Professional Documents
Culture Documents
3Bmt MICR 231 LEC - Bacteriology
Uploaded by
roseannequyo0 ratings0% found this document useful (0 votes)
12 views2 pagesPrinciple of the Urease Test in Bacteriology
Original Title
Urease Test
Copyright
© © All Rights Reserved
Available Formats
DOCX, PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentPrinciple of the Urease Test in Bacteriology
Copyright:
© All Rights Reserved
Available Formats
Download as DOCX, PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
12 views2 pages3Bmt MICR 231 LEC - Bacteriology
Uploaded by
roseannequyoPrinciple of the Urease Test in Bacteriology
Copyright:
© All Rights Reserved
Available Formats
Download as DOCX, PDF, TXT or read online from Scribd
You are on page 1of 2
3BMT MICR 231 LEC Bacteriology
Biochemical Tests for Bacterial Identification
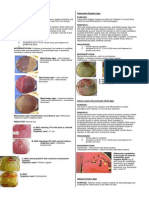
Urease Test (Christensen's Method)
Purpose:
Determination of an organism's ability to
produce urease, an enzyme which hydrolyzes
urea. Presumptive identification of Proteus sp.
may be possible through its ability to rapidly
hydrolyze urea.
Priniciple:
Urea is the product of decarboxylation of amino
acids. Hydrolysis of urea produces ammonia
and CO
2
. The formation of ammonia alkalinizes
the medium, and the pH shift is detected by the
color change of phenol red from light orange at
pH 6.8 to magenta (pink) at pH 8.1. Rapid
urease-positive organisms turn the entire
medium pink within 24 hours. Weakly positive
organisms may take several days, and negative
organisms produce no color change or yellow as
a result of acid production.
Media:
Enzymatic digest of gelatin (1 g), dextrose (1 g),
NaCl (5 g), KH
2
PO
4
(2 g), urea (20 g), phenol red
(0.012 g), per 1000 mL, pH 6.8.
Method:
1. Streak the surface of a urea agar slant with a
portion of a well-isolated colony or inoculate
slant with 1 to 2 drops from an overnight brain-
heart infusion broth.
2. Leave the cap on loosely and incubate the
tube at 35-37C in ambient air for 48 hours to 7
days.
Expected Results:
Positive: Change in color of slant from light
orange to magenta.
Negative: No color change (agar slant and butt
remain light orange)
Limitations:
Alkaline reactions may appear after prolonged
incubation and may be the result of peptone or
other protein utilization raising the pH. To
eliminate false-positive reactions, perform a
control test with the base medium without
urea.
Quality Control:
Positive: Proteus vulgaris (ATCC13315)
Weak positive: Klebsiella pneumoniae
(ATCC13883)
Negative: Escherichia coli (ATCC25922)
You might also like
- Identification of Pseudomonas SPDocument20 pagesIdentification of Pseudomonas SPUttam Kr Patra100% (4)
- Microbiology ReviewerDocument8 pagesMicrobiology ReviewerSteph Vee100% (6)
- Biochemical Test of BacteriaDocument33 pagesBiochemical Test of Bacteriaaziskf100% (2)
- Clinical MicrosDocument28 pagesClinical MicrosKristine Jamella Maris NaragNo ratings yet
- 3 Chemical Examination of UrineDocument82 pages3 Chemical Examination of UrineJake Real Dela RocaNo ratings yet
- UrinalysisDocument43 pagesUrinalysisJames Knowell75% (4)
- Biochemistry Lab Con ProteinsDocument47 pagesBiochemistry Lab Con Proteinsriana santosNo ratings yet
- Biochemistry AnalyzerDocument23 pagesBiochemistry AnalyzerBunga ExsotikaNo ratings yet
- Biochemical Tests For Identification of Bacteria PDFDocument30 pagesBiochemical Tests For Identification of Bacteria PDFEmber BahianNo ratings yet
- Plant and Animal Bio-Chemistry - Including Information on Amino Acids, Proteins, Pigments and Other Chemical Constituents of Organic MatterFrom EverandPlant and Animal Bio-Chemistry - Including Information on Amino Acids, Proteins, Pigments and Other Chemical Constituents of Organic MatterNo ratings yet
- Miscellaneous Biochemical TestsDocument45 pagesMiscellaneous Biochemical TestsDale Daniel Macarandan SisonNo ratings yet
- 1110Document2 pages1110vaibhavNo ratings yet
- Catalog # URS-10: 100 Strips Per BoxDocument4 pagesCatalog # URS-10: 100 Strips Per BoxLey100% (1)
- 9 Lecture 2019-09-26 07 - 48 - 01Document53 pages9 Lecture 2019-09-26 07 - 48 - 01gothai sivapragasamNo ratings yet
- Tanner Scientific 10SG Reagent Test StripsDocument2 pagesTanner Scientific 10SG Reagent Test Stripssaifulmangopo123No ratings yet
- AUBF Lec Week#5 Chemical ExaminationDocument48 pagesAUBF Lec Week#5 Chemical ExaminationLexaNatalieConcepcionJuntadoNo ratings yet
- Lab 8Document28 pagesLab 8Mustafa A. DawoodNo ratings yet
- Biochemical TestsDocument6 pagesBiochemical TestsCherie QuintoNo ratings yet
- Multistix Reagent StripDocument3 pagesMultistix Reagent StripFrancis TorresNo ratings yet
- Biochem TestsDocument27 pagesBiochem TestsSteph VeeNo ratings yet
- Biochemical TestsDocument41 pagesBiochemical TestsChinthikaNo ratings yet
- Group 2 - Bacte Lab. ExamDocument11 pagesGroup 2 - Bacte Lab. ExamANo ratings yet
- Us Vis13Document2 pagesUs Vis13Nghi NguyenNo ratings yet
- Microbiolal TestingDocument8 pagesMicrobiolal Testingmalakst200No ratings yet
- Biochemical-Tests 1Document7 pagesBiochemical-Tests 1Charlyn Dela CruzNo ratings yet
- Standrerds For DNS Method ProtocolDocument27 pagesStandrerds For DNS Method ProtocolvennellaNo ratings yet
- Urine StripsDocument4 pagesUrine StripsABUAC CYSSELLE DAWN KNo ratings yet
- UROBILINOGEN (60 Seconds)Document5 pagesUROBILINOGEN (60 Seconds)Margaret Nicole AboNo ratings yet
- Lec 3 BacteDocument7 pagesLec 3 BacteMitchee ZialcitaNo ratings yet
- Biochemical Tests IMViCDocument3 pagesBiochemical Tests IMViCSayyeda SumaiyahNo ratings yet
- 1062Document2 pages1062Nisadiyah Faridatus ShahihNo ratings yet
- CAT Nº: 1208: Lysine Decarboxylase BrothDocument2 pagesCAT Nº: 1208: Lysine Decarboxylase BrothNguyễnVinhNo ratings yet
- Urease Test - Principle, Media, Procedure and ResultDocument11 pagesUrease Test - Principle, Media, Procedure and ResultAlisha KhanNo ratings yet
- 201020120500-CLED Agar With Andrade IndicatorDocument3 pages201020120500-CLED Agar With Andrade Indicatorsofimar sanzNo ratings yet
- IMViC TestsDocument3 pagesIMViC Testsdr.pragna infoplusNo ratings yet
- CAT Nº: 1405: Saline Peptone Water Iso 6887Document2 pagesCAT Nº: 1405: Saline Peptone Water Iso 6887Bldjl HichemNo ratings yet
- Identification of Staphylococcus Species Using API Staph Commercial KitDocument8 pagesIdentification of Staphylococcus Species Using API Staph Commercial KitCherisse TuazonNo ratings yet
- Lab Policies Urinalysis - Clinetek Status and Visual Lab 1583Document17 pagesLab Policies Urinalysis - Clinetek Status and Visual Lab 1583hunnylandNo ratings yet
- Indole TestDocument3 pagesIndole TestNisha HernandezNo ratings yet
- Screenshot 2023-03-28 at 11.36.42 AMDocument48 pagesScreenshot 2023-03-28 at 11.36.42 AMSourayaNo ratings yet
- Urinalysis Materials and Equipment, MethodsDocument7 pagesUrinalysis Materials and Equipment, MethodsAlanah JaneNo ratings yet
- UrinalysisDocument67 pagesUrinalysisumairzafar5261No ratings yet
- Tests For Glucose 1 - Benedict'S Test: Cus04 (Cupric Sulfide) + Reducing Substance HeatDocument17 pagesTests For Glucose 1 - Benedict'S Test: Cus04 (Cupric Sulfide) + Reducing Substance HeatLyka PapaNo ratings yet
- Antibiotic Senstivity of Various Pathogenic Bacteria in Case of Uti and Throat InfectionsDocument25 pagesAntibiotic Senstivity of Various Pathogenic Bacteria in Case of Uti and Throat InfectionsMishel AroraNo ratings yet
- Media Biochemical TestsDocument36 pagesMedia Biochemical TestsIsmail Bazly ZarirNo ratings yet
- 005-Clinical MicroDocument15 pages005-Clinical Micro04051914001No ratings yet
- Media & Biochemical Tests: Laboratory ObjectivesDocument36 pagesMedia & Biochemical Tests: Laboratory ObjectivesHiba Abu-jumahNo ratings yet
- Biochemistry Lab ReviewDocument5 pagesBiochemistry Lab ReviewFemi BeautyNo ratings yet
- Uses of Carbohydrate Fermentation Test: EnterobacteriaceaeDocument12 pagesUses of Carbohydrate Fermentation Test: EnterobacteriaceaeChristyl JoNo ratings yet
- Upload 00473492 1531131869366Document5 pagesUpload 00473492 1531131869366Nhan DoNo ratings yet
- Violet Red Bile Glucose Agar (V.R.B.G.A.) : Enterobacteriaceae in Foodstuffs With MPN Technique WithoutDocument7 pagesViolet Red Bile Glucose Agar (V.R.B.G.A.) : Enterobacteriaceae in Foodstuffs With MPN Technique WithoutThandie KehleNo ratings yet
- Biochemical TestDocument25 pagesBiochemical TestBiswajit DharuaNo ratings yet
- Biochemical Test Final-CH 6Document40 pagesBiochemical Test Final-CH 6SisayNo ratings yet
- Order of Biochemical TestsDocument73 pagesOrder of Biochemical Testskenny1973No ratings yet
- Urine TestDocument104 pagesUrine Testdustpops100% (1)
- Catalase TestDocument12 pagesCatalase Testshiva121294No ratings yet
- FPBB Laboratory ManualDocument26 pagesFPBB Laboratory ManualRock StarNo ratings yet
- Oligonucleotide-Based Drugs and Therapeutics: Preclinical and Clinical Considerations for DevelopmentFrom EverandOligonucleotide-Based Drugs and Therapeutics: Preclinical and Clinical Considerations for DevelopmentNicolay FerrariNo ratings yet