Professional Documents
Culture Documents
Prebiotic Multi-Letter Polymerization Networks With Chiral Symmetry Breaking
Uploaded by
Damian SowinskiOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Prebiotic Multi-Letter Polymerization Networks With Chiral Symmetry Breaking
Uploaded by
Damian SowinskiCopyright:
Available Formats
Prebiotic Multi-letter Polymerization Networks with Chiral Symmetry Breaking
Damian R. Sowinski, Department of Physics and Astronomy, Dartmouth College
The Model
We begin with an alphabet of N monomers:
Motivation
Living organisms on Earth, without exception, utilize chiral molecules within their metabolic networks. All sugars are right handed, dextromers, while chiral amino acids are left handed, levomers. Laboratory synthesis of both sugars and amino acids result in racemic solutions. This poses a very interesting problem for those delving into the most profound question of the Origin of Life. How and when did the enantiomeric excess in these biomolecules emerge from the natural conditions that existed in the prebiotic soup of the early Earth? In this work we examine a simple mathematical model of a polymerization network consisting of a multi-letter chiral monomer alphabet. Chiral and achiral monomer coupling dependent rate constants are considered, as is the delity of substrate to monomer generation and an explicit bias. The strength of the general multi-letter alphabet rests in its ability to describe chiral symmetry breaking in a wide range of systems. 1-letter alphabets: polymerization networks of sugars, such as glucose. 2-letter alphabets: chiral information strings, sugar-amino acid interaction networks 4-letter alphabets: RNA and DNA synthesis 20+ letter alphabets: polymerization of amino acids into unfolded proteins An explicit bias and delity can be put in by hand, though spontaneous symmetry breaking does occur for large segments of the parameter space of monomer couplings. Surprisingly, the effective potential the asymmetry eld lives in is not very sensitive to many parameters. We begin our analysis by examining a simplied model at the dimer level, under the adiabatic approximation wherein the substrate is kept at a constant amount and higher level polymer growth in negligible.
Future Directions
The next part of this work will go into examining the parameter space of the 2-letter alphabet, to determine the range of coupling parameters that are conducive to symmetry breaking. Moving beyond the dimer model will allow a more thorough exploration of the sequence space population as well. Spatial dependence will be introduced via a diffusion term with a polymer length dependent diffusion constant. Hydrolysis cycles will also be easy to incorporate by allowing for backow between polymer spaces. Finally, Universal Template Replication will be incorporated to begin modeling RNA and DNA. The future is bright with possibilities!
A = {A1 , A2 , . . . AN }
The sequence space is constructed by taking the union of quotient spaces of the m-letter primitive word lattices with respect to the equivalence relation generated by the reversal operator: 0 =w : A m Am : w ~ 7 w ~ 0 | wn mn
w ~1 w ~2 w ~ 1 = ( w ~ 2)
Wm Am =
S=
m=1
Wm
Due to the cross-inhibition of polymer growth, chirality is incorporated into the model by introducing four polymer families: 1. The homochiral polymers
Source
AC n =
C {A~ v
|~ v Wn , Chirality (Avi ) = C i}
AC 1
AC 1
2. The left-inhibited polymers ( n > 2 )
C C Bn = {B~ | ~ v W , Chirality ( A ) = C, n v 1 v
The last bits needed to describe the dynamics are the source equations for the monomers. To simplify the analysis, we made the sources have the same form for all the monomers of the same chirality. The delity, f, controls the accuracy with which the polymers of one chirality induce the production of same-chirality monomers. 1 c o r r e s p o n d t o p e r f e c t d e l i t y, w h e r e dextropolymers make only dextromonomers, and levopolymers make only levomonomers. 0 corresponds to each polymer making 50/50 of each type of monomer. The bias, g, determines how effective dextro versus levo monomer production is. 0 corresponds to perfect symmetry between dextro and levo production. Positive bias is in the levo direction.
Adiabatic Approximation and the Reduced Bias Model, N=1
In the adiabatic approximation we set the growth of polymers longer than the dimer level equal to 0. We examine the case of a one letter alphabet, and nd generalized results for the model developed by Gleiser and Walker. The symmetry (asymmetry) elds are dened by taking the sum (difference) of chiral concentrations. We nd that the asymmetry eld lives in an effective potential with spontaneous symmetry breaking when the symmetry eld is in equilibrium. Increasing the heterochiral chiral coupling, we nd the effective potential wells are pushed upward, eventually destroying the possibility of chiral symmetry breaking when bias is nonexistent.
Chirality (Avi ) = C i 6= 1}
AC 2
D2
AC 2
3. The right-inhibited polymers ( n > 2 )
C Cn
C {C~ v
|~ v Wn , Chirality (Avn ) = C,
C B3
AC 3
C C3
C C3
AC 3
C B3
Chirality (Avi ) = C i 6= n}
C D4 C B4
AC 4
C C4
C C4
C A4
C B4
C D4
4. The frozen polymers ( n = 2, n > 3 )
C Dn
C {D~ v
|~ v Wn , Chirality (Av1 ) = C,
Chirality (Avn ) = C,
g=0.1
Asymmetry Field
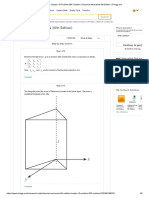
The polymerization network can be visualized via a directed graph, with polymers in the above dened spaces getting mapped to higher sequence length spaces as they absorb monomers. Here the solid arrows represent homochiral polymerization, whereas dashed arrows represent heterochiral growth. The boxed regions show us that the rate reaction equations for the monomers, dimers, and trimers will be unique. All higher polymers will have similar evolution equations, all of which can be read off from the diagram.
o X X n d [ AC ] C C C i C C C C (i,s1 + i,s2 )[A~ (1 + i,s )i,s [As ] + i,s [As ] [Ai ] = Q i [ Ai ] s ] s ] + (i,s1 + i,s2 )[A~ dt ~ s W2 s W1 X X C C C C C C C (i,s1 + i,sn )[A~ s ] + i,sn [B~ s ] + i,sn [A~ s ] + i,s1 [C~ s ] + i,sn [B~ s ] + i,s1 [C~ s ] s ] + i,s1 [A~
n=3 ~ s Wn
Asymmetry Field
Chirality (Avi ) = C i 6= 1, n}
0.2 0.4 0.0 -0.2 -2 -1 0.0 0.2
-1
-1
C d[A~ C C C C C s ] = s1 ,s2 [AC ][ A ] | +( [ A ][ A ] + [ A ][ A ~ s W sWn ,n>2 s1 ,1 (~ s)1 s2 ,2 (~ s)2 s1 s2 s1 s2 2 1 (~ s) 2 (~ s) ]) |~ dt
-2 0.0 0.5 1.0 1.5 2.0
-2 0.0 0.5 1.0 1.5 2.0
-0.2 1 2 -0.4
Symmetry Field f=1, g=0, e=0
Symmetry Field f=1, g=.1, e =0
2
s0 W1
C C C 0 ,s + s0 ,s )[A 0 ][A ] ][ A ] + ( (s0 ,s1 + s0 ,s2 )[AC 0 s s s 2 1 ~ s ~ s
1
1
Asymmetry Field
Asymmetry Field
C X d[B~ ] C C C C C C C C s 0 ,s [B ][A 0 ] + s0 ,s [B ][A 0 ] ][ A ] | ] + [ B = s1 ,1 (~ s ~ s W ,n> 2 s)1 [As1 ][A1 (~ s , ( ~ s ) s s s 2 2 ~ s ~ s n s) 2 (~ s) 2 2 2 2 dt 0
s W1
g=0.2
0.2
AC i
C Ai
+
+
AC i +
AC i
C iv1 A~ v ivn C iv1 A~ v ivn C ivn B~ v
C ivn B~ v
C X d[C~ ] C C C C C C C s s0 ,s1 [AC = s2 ,2 (~ sWn ,n>2 s)2 [A2 (~ s)1 [As1 ][C1 (~ s0 ][C~ s ] + s0 ,s1 [As0 ][C~ s ] s) ][As2 ] + s1 ,1 (~ s) ] |~ dt 0
0.1
AC (i,~ v) ,
C B( i,~ v) , C B( ~ v ,i)
C D( ~ v ,i)
AC (~ v ,i)
C C( ~ v ,i)
QC i
g 1+f 1f C = Q = kC [S ](1 + C )( PC + PC ), PC = PC ([A~ s ]) 2 2 2
C
s W1
-1
-1
-2
-1
-2
-2
0.0
0.5
1.0
1.5
2.0
0.0 0.5 1.0 1.5 2.0
References
[1] ! Gleiser, Marcelo, and Sara I. Walker. "An Extended Model for the Evolution of Prebiotic Homochirality: A Bottom-Up Approach to the Origin ! of Life." ArXiv:0802.2884 (n.d.). [2] ! Gleiser, Marcelo, and Sara I. Walker. "The Chirality Of Life: From Phase Transitions To Astrobiology." ArXiv:0811.1291 (n.d.) [3] ! Gleiser, Marcelo, Bradley J. Nelson, and Sara I. Walker. "Chiral Polymerization in Open Systems From Chiral-Selective Reaction Rates." ArXiv: 1112.1393 (n.d.) [4] ! Gleiser, Marcelo. "Asymmetric Spatiotemporal Evolution of Prebiotic Homochirality." ArXiv:astro-ph/0606593 (n.d.) [5] ! Cronin, John R., and Sandra Pizzarello. "Enantiomeric Excesses in Meteoritic Amino Acids." Science. AAAS, n.d. Web. <http:// www.sciencemag.org/content/275/5302/951.short>.
Symmetry Field f=1, g=0, e=-1
Symmetry Field f=1, g=.1, e=.5
-0.1
AC i
C Ai
+
+
C iv1 C~ v C iv1 C~ v
C C( i,~ v)
C D( i,~ v)
The effects of indelity are to push the true vacua closer to one another, hence leading to an incomplete symmetry breaking event. It is surprising to see that all it takes is just a very small bias to pusht he system completely into a single chiral state.
-0.2
You might also like
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5795)
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (588)
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (74)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (895)
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (838)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (400)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (345)
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2259)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (266)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1090)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (121)
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- ATOMEX Europe 2011: Sigma Group A.SDocument35 pagesATOMEX Europe 2011: Sigma Group A.StomognNo ratings yet
- CSEC Mathematics - EvaluationDocument5 pagesCSEC Mathematics - EvaluationAnthony BensonNo ratings yet
- Panasonic TX L32C10P 10PS Chassis GLP24Document38 pagesPanasonic TX L32C10P 10PS Chassis GLP24yacosNo ratings yet
- I RL 3010.0F 1350 960 PPC 004 - CDocument32 pagesI RL 3010.0F 1350 960 PPC 004 - ChadrijkNo ratings yet
- Submitted Lead PollutionDocument4 pagesSubmitted Lead PollutionRakesh JainNo ratings yet
- Career Point: Fresher Course For IIT JEE (Main & Advanced) - 2017Document2 pagesCareer Point: Fresher Course For IIT JEE (Main & Advanced) - 2017kondavetiprasadNo ratings yet
- Lce UserDocument34 pagesLce Userapi-3831923No ratings yet
- Solved - Chapter 10 Problem 28P Solution - Classical Mechanics 0th EditionDocument6 pagesSolved - Chapter 10 Problem 28P Solution - Classical Mechanics 0th EditionRyuNo ratings yet
- CH4 - Jan 2014Document15 pagesCH4 - Jan 2014Kieran RichardsNo ratings yet
- Practice Projectiles 2Document4 pagesPractice Projectiles 2fatpelicanNo ratings yet
- Manual Daikin VRVDocument20 pagesManual Daikin VRVErnesto BonillaNo ratings yet
- Red TactonDocument15 pagesRed TactonAbhijeet ChattarjeeNo ratings yet
- Load CalculationDocument4 pagesLoad CalculationMalayKumarDebNo ratings yet
- ICP OES Maintenance Troubleshooting PDFDocument39 pagesICP OES Maintenance Troubleshooting PDFmiftahchemNo ratings yet
- ControlLogix® 5560M03SE Combination Controller and SERCOS Interface - 1756-In593a-En-PDocument48 pagesControlLogix® 5560M03SE Combination Controller and SERCOS Interface - 1756-In593a-En-PElder MartinsNo ratings yet
- 10-1 PaintDocument5 pages10-1 PaintdetteheartsNo ratings yet
- 4NA - Heat TransferDocument24 pages4NA - Heat TransferChico AlvesNo ratings yet
- ASTM E6-23aDocument12 pagesASTM E6-23aHunter 911No ratings yet
- Introduction To Magnetism and Magnetic Materials: David JilesDocument5 pagesIntroduction To Magnetism and Magnetic Materials: David JilesAmreshAman0% (1)
- Mechanical Engineering - Final PDFDocument248 pagesMechanical Engineering - Final PDFRubal SharmaNo ratings yet
- Elg4152 Modern Control Engineering Textbook Modern 59ce3e8c1723dd6dc1b864beDocument1 pageElg4152 Modern Control Engineering Textbook Modern 59ce3e8c1723dd6dc1b864bedhapraNo ratings yet
- Allied 3 - Mathematical Statistics I - Semester IIIDocument3 pagesAllied 3 - Mathematical Statistics I - Semester IIISwethaNo ratings yet
- Theoretical Optics: Pre Pared By: Je Nno S - La Ntaya, OdDocument27 pagesTheoretical Optics: Pre Pared By: Je Nno S - La Ntaya, OdDyra Kim QuintanaNo ratings yet
- Problem 12-105Document53 pagesProblem 12-105adam johnsonNo ratings yet
- United State Pharmacopoea 1st EditionDocument294 pagesUnited State Pharmacopoea 1st EditionNehruRogerNo ratings yet
- Book 1 Electronic Devices and Circuit ApplicationsDocument319 pagesBook 1 Electronic Devices and Circuit ApplicationsRoze100% (1)
- Lab Report: Subject: Chemical Reaction EngineeringDocument56 pagesLab Report: Subject: Chemical Reaction EngineeringAbdul RehmanNo ratings yet
- Rsgis - .Module I - FundamentalsDocument25 pagesRsgis - .Module I - FundamentalsnambimunnaNo ratings yet
- Solve Study Textbooks: Open in AppDocument4 pagesSolve Study Textbooks: Open in AppAdi PNo ratings yet
- RESUME Marjan Shameem QTRDocument5 pagesRESUME Marjan Shameem QTRsophist1No ratings yet