Professional Documents
Culture Documents
FTIR Pectra PDF
Uploaded by
Vinícius PiantaOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
FTIR Pectra PDF
Uploaded by
Vinícius PiantaCopyright:
Available Formats
Spectroscopy Data Tables 1
Infrared Tables (short summary of common absorption frequencies)
The values given in the tables that follow are typical values. Specific bands may fall over a range of
wavenumbers, cm-1. Specific substituents may cause variations in absorption frequencies. Absorption
intensities may be stronger or weaker than expected, often depending on dipole moments. Additional
bands may confuse the interpretation. In very symmetrical compounds there may be fewer than the
expected number of absorption bands (it is even possible that all bands of a functional group may
disappear, i.e. a symmetrically substituted alkyne!). Infrared spectra are generally informative about what
functional groups are present, but not always. The 1H and 13C NMRs are often just as informative about
functional groups, and sometimes even more so in this regard. Information obtained from one
spectroscopic technique should be verified or expanded by consulting the other spectroscopic techniques.
IR Summary - All numerical values in the tables below are given in wavenumbers, cm-1
Bonds to Carbon (stretching wave numbers)
sp3 C-X single bonds sp2 C-X single bonds
C N C O C O
C C C C C N
1000-1350 1050-1150 1100-1350
not used not very useful alkoxy C-O not very useful 1250 acyl and phenyl C-O
sp2 C-X double bonds sp C-X triple bonds
C O
C C C N
C C C N
1640-1810
expanded table
1600-1680 1640-1690 on next page 2100-2250 2240-2260
Stronger dipoles produce more intense IR bands and weaker dipoles produce less intense IR bands (sometimes none).
Bonds to Hydrogen (stretching wave numbers)
C H
O
C C H
C H C H
3000-3100
sp3 C-H 3300 2700-2760
sp3 C-H 2800-2860
2850-3000 (see sp2 C-H bend aldehyde C-H
sp3 C-H patterns below) (sp C-H bend 620) (two bands)
H
O
C N C N R S H
R O H C O H
H H
3100-3500 3100-3500 2550 -2620
primary NH2 secondary N-H 3200-3400 2500-3400 (very weak)
(two bands) (one band)
alcohol O-H acid O-H thiol S-H
amides = strong, amines = weak
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 2
Carbonyl Highlights (stretching wave numbers)
Aldehydes Ketones Esters Acids
O O O O
C C C R' C H
R H R R R O R O
saturated = 1725 saturated = 1715 saturated = 1735 saturated = 1715
conjugated = 1690 conjugated = 1680 conjugated = 1720 conjugated = 1690
aromatic = 1700 aromatic = 1690 aromatic = 1720 aromatic = 1690
6 atom ring = 1715 6 atom ring = 1735
5 atom ring = 1745 5 atom ring = 1775
4 atom ring = 1780 4 atom ring = 1840
3 atom ring = 1850
Amides Anhydrides Acid Chlorides nitro
O O O
O
O
C C
C R N
R O R R Cl
R NR2
saturated = 1650 saturated = 1760, 1820 saturated = 1800 O
conjugated = 1660 conjugated = 1725, 1785 conjugated = 1770
aromatic = 1725, 1785 aromatic = 1770 asymmetric = 1500-1600
aromatic = 1660 symmetric = 1300-1390
6 atom ring = 1670 6 atom ring = 1750, 1800
5 atom ring = 1700 5 atom ring = 1785, 1865
4 atom ring = 1745 Very often there is a very weak C=O overtone at approximately 2 x (3400 cm-1).
3 atom ring = 1850 Sometimes this is mistaken for an OH or NH peak.,
sp2 C-H bend patterns for alkenes sp2 C-H bend patterns for aromatics
absorption absorption
alkene substitution descriptive frequencies (cm-1) aromatic substitution descriptive frequencies (cm-1)
pattern alkene term pattern aromatic term due to sp2 CH bend
due to sp2 CH bend
R H
monosubstituted 985-1000 monosubstituted 690-710
C C X
alkene 900-920 aromatic 730-770
H H
R R X
C C cis disubstituted 675-730
alkene (broad) 735-770
ortho disubstituted
H H X aromatic
R H
C C trans disubstituted 960-990
alkene X
H R
R H
meta disubstituted 680-725
X
geminal disubstituted 880-900 aromatic 750-810
C C
alkene 880-900 (sometimes)
R H
R R
trisubstituted para disubstituted 790-840
C C 790-840 X X aromatic
alkene
R H
R R Aromatic compounds have characteristic weak overtone bands
that show up between 1650-2000 cm-1). Some books provide
C C tetrasubstituted
alkene none pictures for comparison (not here). A strong C=O peak will
R R cover up most of this region.
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 3
units = cm-1
4000 3500 3000 2500 2000 1700 1500 1400 1300 1200 1100 1000 900 800 700 600 500
sp3 C-H C C C=O alkene sp2 C-H
sp C-H bend
stretch stretch stretch 3
sp C-H
thiol S-H bend mono
sp2 C-H stretch
C=N cis
stretch C N stretch trans
aldehyde C-H acyl C-O geminal
stretch phenol C-O
C=C stretch tri
alcohol O-H alkene
stretch
alkoxy C-O
carboxylic acid O-H C=C stretch
aromatic aromatic sp2 C-H
stretch
bend
1o N-H2 mono
N-H bend
stretch
ortho
2o N-H nitro nitro meta
stretch para
4000 3500 3000 2500 2000 1700 1500 1400 1300 1200 1100 1000 900 800 700 600 500
expansion of alkene & aromatic sp2 C-H bend region (units = cm-1)
1000 900 800 700 600 500
mono mono
cis
alkene sp2 C-H
trans
geminal bend
tri
mono mono
ortho aromatic sp2 C-H
bend
meta meta meta
para
expansion of carbonyl (C=O) stretch region (units = cm-1)
1800 1750 1700 1650 1600
Saturated C=O lies at carboxylic acid C=O (also acid "OH") Conjugated C=O
higher cm-1 lies at lower cm-1
C=O in samll rings ester C=O (also acyl C-O and alkoxy C-O)
lies at higher cm-1
aldehyde C=O (also aldehyde C-H)
ketone C=O (nothing special)
amide C=O (low C=O, amide N-H)
acid chloride C=O (high C=O, 1 peak)
anhydride C=O anhydride C=O (high C=O, 2 peaks)
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 4
IR Flowchart to determine functional groups in a compound (all values in cm-1).
IR Spectrum
has C=O band
(1650-1800 cm-1) does not have
C=O band
very strong C C
C N
aldehydes
O alkanes
1725-1740 (saturated) 2250 sp3 C-H stretch 2850-3000
1660-1700 (unsaturated) nitriles
C sharp, stronger 1460 & 1380
sometimes lost than alkynes, sp3 C-H bend
2860-2800 in sp3 CH peaks C N
C C not useful
aldehyde C-H 2760-2700 a little lower
(both weak) when conjugated alkenes
sp2 C-H stretch 3000-3100
ketones alkynes 650-1000
O 2150 sp2 C-H bend (see table for
1710-1720 (saturated) C C spectral patterns)
1680-1700 (unsaturated) (variable intensity)
C 1715-1810 (rings: higher not present or weak when symmetrically 1600-1660
in small rings) substituted, a little lower when conjugated C C
weak or not present
esters - rule of 3
O sp C-H stretch 3300 aromatics
1735-1750 (saturated) sharp, strong
1715-1740 (unsaturated) sp2 C-H stretch 3050-3150
C 1735-1820 (higher in small rings) sp C-H bend 620 690-900 (see table),
acyl sp2 C-H bend overtone patterns
1150-1350 (acyl, strong) All IR values are approximate and have a range between 1660-2000
C O
alkoxy of possibilities depending on the molecular 1600 & 1480
C O
(1000-1150, alkoxy, medium) environment in which the functional group resides. C C can be weak
Resonance often modifies a peak's position
acids because of electron delocalization (C=O lower,
O alcohols
1700-1730 (saturated) acyl C-O higher, etc.). IR peaks are not 100%
reliable. Peaks tend to be stronger (more intense) alcohol
1715-1740 (unsaturated) 3600-3500
C when there is a large dipole associated with a O H
1680-1700 (higher in small rings)
vibration in the functional group and weaker in alkoxy 1000-1260
acyl less polar bonds (to the point of disappearing in C O (3o > 2o > 1o)
C O 1210-1320 (acyl, strong) some completely symmetrical bonds).
thiols
acid 2550 (weak)
2400-3400, very broad thiol
O H (overlaps C-H stretch) 2 S H (easy to overlook)
Alkene sp C-H bending patterns
amides amines
O monosubstituted alkene (985-1000, 900-920) H
3300 - 3500, two bands
1630-1680 (saturated) geminal disubstituted (960-990)
1745 (in 4 atom ring) N for 1o amines, one band
C cis disubstituted (675-730) o
N H for 2 amines, weaker
trans disubstituted (880-900) 1o
H 3350 & 3180, two bands H
2o than in amides,
trisubstituted (790-840)
for 1o amides,
tetrasubstituted (none, no sp2 C-H) N-H bend, 1550-1640,
N one band for 2o amides, N H
stronger in amides than amines
o
1 N H stronger than in amines, extra
H
2o overtone sometimes at 3100 N C 1000-1350
Aromatic sp2 C-H bending patterns (uncertain)
ethers
N H N-H bend, 1550-1640, monosubstituted (730-770, 690-710) 1120 (alphatic)
stronger in amides than amines alkoxy
ortho disubstituted (735-770) C O 1040 & 1250 (aromatic)
acid chlorides meta disubstituted (880-900,sometimes,
nitro compounds
O 1800 (saturated) 750-810, 680-725)
O
1770 (unsaturated) para disubstituted (790-840) 1500-1600, asymmetric (strong)
C Inductive pull of Cl increases the N 1300-1390, symmetric (medium)
electron density between C and O. O
There are also weak overtone bands between
anhydrides 1660 and 2000, but are not shown here. You
O can consult pictures of typical patterns in other carbon-halogen bonds
1760 & 1820 (saturated) reference books. If there is a strong C=O band,
1725-1785 (unsaturated) they may be partially covered up.
C two strong bands
C X usually not
acyl very useful
C O 1150-1350 (acyl, strong)
X = F, Cl, Br, I
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 5
deshielding side = less electron rich Typical 1H and 13C NMR chemical shift values. shielding side = more electron rich
(inductive & resonance) (inductive & resonance)
typical proton chemical shifts
amine N-H
Carbon and/or heteroatoms without hydrogen do
not appear here, but influence on any nearby protons 2 1
may be seen in the chemical shifts of the protons. alcohol O H
5 1
amide N-H
6 1
S C H
thiols, sulfides
2.5 2.0
N C H
amines
X C H
3.0 2.3
X = F,Cl,Br,I allylic C-H
5 3 2.5 1.5
benzylic C-H thiol
alkene C-H carbonyl alpha C-H SH
carboxylic acid O-H 7+ 4 3+ 2 1.5 1.3
epoxide C-H
12 10
3.5 2.5
H
alcohols simple sp3 C-H
O C ethers
aldehyde C-H aromatic C-H CH > CH2 > CH3
esters C H
10 9 8+ 6 5+ 3.3 3 2 2 0.5
12 11 10 9 8 7 PPM 6 5 4 3 2 1 0
typical carbon-13 chemical shifts F 80-95
halogen C
Cl 45-70
Br 35-65
with & without H I 15-45
O
95 15
C N C
R R
ketones
amines, amides
no H with & without H
220+ 180 50 30
O
O
C
R X epoxides
C C with & without H
carboxylic acids
anhydrides with & without H
N C 60 40
esters
amides no H 90 +
70 -
S C
acid chlorides
125 110 thiols, sulfides
no H
with & without H
180 160 O C alcohols, 40 20
ethers, esters
O
with & without H
C +
R H C 80 50
C C simple sp3 carbon
aldehydes C > CH > CH2 > CH3
with H with & without H with & without H
210 180 160+ 100- 60+ 0
240 220 200 180 160 140 PPM 120 100 80 60 40 20 0
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 6
Calculation of chemical shifts for protons at sp3 carbons
Estimation of sp3 C-H chemical shifts with multiple substituent parameters for protons within 3 C's of consideration.
H
C C C = directly attached substituent, use these values when the hydrogen and substituent are attached to the same carbon
= once removed substituent, use these values when the hydrogen and substituent are on adjacent (vicinal) carbons
= twice removed substituent, use these values when the hydrogen and substituent have a 1,3 substitution pattern
Starting value and equations for CH3's
X = substituent
R- (alkyl) 0.0 0.0 0.0
R2C=CR- (alkenyl) 0.8 0.2 0.1 CH3 = 0.9 + H3C
RCC- (alkynyl) 0.9 0.3 0.1
Ar- (aromatic) 1.4 0.4 0.1
F- (fluoro) 3.2 0.5 0.2 CH3 = 0.9 + ( + ) H3C C C
Cl- (chloro) 2.2 0.5 0.2
Br- (bromo) 2.1 0.7 0.2
I- (iodo) 2.0 0.9 0.1
is the summation symbol for all substituents considered
HO- (alcohol) 2.3 0.3 0.1
RO- (ether) 2.1 0.3 0.1
epoxide 1.5 0.4 0.1 Starting value and equation for CH2's
R2C=CRO- (alkenyl ether) 2.5 0.4 0.2
ArO- (aromatic ether) 2.8 0.5 0.3 In a similar manner we can calculate chemical shifts
RCO2- (ester, oxygen side) 2.8 0.5 0.1 for methylenes (CH2) using the following formula
ArCO2- (aromatic ester, oxygen side) 3.1 0.5 0.2
ArSO3- (aromatic sulfonate, oxygen) 2.8 0.4 0.0 CH2 = 1.2 + ( + + ) H
H2N- (amine nitrogen) 1.5 0.2 0.1 H C C C
RCONH- (amide nitrogen) 2.1 0.3 0.1
O2N- (nitro) 3.2 0.8 0.1
is the summation symbol for all substituents considered
HS- (thiol, sulfur) 1.3 0.4 0.1
RS- (sulfide, sulfur) 1.3 0.4 0.1 Starting value and equation for CH's
OHC- (aldehyde) 1.1 0.4 0.1
RCO- (ketone) 1.2 0.3 0.0
ArCO- (aromatic ketone) 1.7 0.3 0.1 In a similar manner we can calculate chemical shifts
HO2C- (carboxylic acid) 1.1 0.3 0.1 for methines (CH) using the following formula
RO2C- (ester, carbon side) 1.1 0.3 0.1 H
CH = 1.5 + ( + + )
H2NOC- (amide, carbon side) 1.0 0.3 0.1
C C C
ClOC- (acid chloride) 1.8 0.4 0.1
NC- (nitrile) 1.1 0.4 0.2
RSO- (sulfoxide) 1.6 0.5 0.3 is the summation symbol for all substituents considered
RSO2- (sulfone) 1.8 0.5 0.3
a. methine b. methylene
Calculations are generally close to
CH2 CH3 actual chemical shifts for a single
d. methyl HO
O CH N substituent, but are less reliable as
the number of substituent factors
H2C H goes up. Multiple substituent factors
H3C H2C c. methyl
tend to overestimate an actual chemical
shift.
e. methylene
f. methylene
a. methine = 1.5 + (1.4) + (2.3) + (0.2) = 5.4 ppm d. methyl = 0.9 + (0.1) = 1.0 ppm
actual = 5.2 actual = 1.0
b. methylene = 1.2 + (1.5) + (0.4) + (0.3) = 3.4 ppm e. methylene = 1.2 + (0.3) = 1.5 ppm
actual = 3.0 and 3.2 actual = 1.7
c. methyl = 0.9 + (1.5) = 2.4 ppm f. methylene = 1.2. + (1.7) = 2.9 ppm
actual = 2.6 actual = 2.9
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 7
Estimated chemical shifts for protons at alkene sp2 carbons
Substituent geminal cis trans Substitution relative to calculated "H"
cis
H- 0.0 0.0 0.0 H
Hydrogen C C
R- 0.5 -0.2 -0.3
trans gem
Alkyl
C6H5CH 2- 0.7 -0.2 -0.2 (ppm) = 5.2 + gem + cis + trans
Benzyl
X-CH2- 0.7 0.1 0.0
Halomethyl Example Calculation
(H)/ROCH2- 0.6 0.0 0.0
alkoxymethyl gem
(H)2/R2NCH2- 0.6 -0.1 -0.1 H
aminomethyl trans
C H
RCOCH2- 0.7 -0.1 -0.1 C
-keto H
NCCH2- 0.7 -0.1 -0.1 CH3O cis
-cyano
R2C=CR- 1.2 0.0 0.0
Alkenyl gem = 5.2 + 1.4 = 6.6
C6H5- 1.4 0.4 -0.1 actual = 6.6
Phenyl
F- 1.5 -0.4 -1.0 trans = 5.2 - 0.1 = 5.1
Fluoro actual = 5.1
Cl- 1.1 0.2 0.1
cis = 5.2 + 0.4 = 5.7
Chloro
actual = 5.6
Br- 1.1 0.4 0.6
Bromo
I- 1.1 0.8 0.9 c d e
Iodo bH H H H
RO- 1.2 -1.1 -1.2 C C C C
akoxy (ether) aH Hf
O C
RCO2- 2.1 -0.4 -0.6
O-ester O
(H)2/R2N- 0.8 -1.3 -1.2 a = 5.2 + (-0.4) = 4.8
N-amino actual = 4.9 (J = 14, 1.6 Hz)
RCONH- 2.1 -0.6 -0.7
N-amide b = 5.2 + (-0.6) = 4.6
O2N- 1.9 1.3 0.6 actual = 4.6 (J = 6, 1.6 Hz)
Nitro c = 5.2 + 2.1 = 7.3
RS- 1.1 -0.3 -0.1 actual = 7.4 (J = 14, 6 Hz)
Thiol
OHC- 1.0 1.0 1.2 d = 5.2 + 0.8 = 6.0
Aldehyde actual = 6.2 (J = 18, 11 Hz)
ROC- 1.1 0.9 0.7 e = 5.2 + 0.5 = 5.7
Ketone actual = 5.8 (J = 11, 1.4 Hz)
HO2C- 0.8 1.0. 03
C-acid f = 5.2 + 1.0 = 6.2
RO2C- 0.8 1.0 0.5 actual = 6.4 (J = 18, 1.4 Hz)
C-ester
H2NOC- 0.4 1.0 0.5
C-amide
NC- 0.3 0.8 0.6
Nitrile
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 8
Estimated chemical shifts for protons at aromatic sp2 carbons
Substitution relative to calculated "H"
Substituent ortho meta para
meta ortho
H- 0.0 0.0 0.0
Hydrogen
CH3- -0.2 -0.1 -0.2 para H
Methyl
ClCH2- 0.0 0.0 0.0 meta ortho
Cholromethyl
Cl3C- 0.6 0.1 0.1 (ppm) = 7.3 + ortho + meta + para
Halomethyl
HOCH 2- -0.1 -0.1 -0.1
Hydroxymethyl
R2C=CR- 0.1 0.0 -0.1 Example Calculation
Alkenyl 2
C6H5- 1.4 0.4 -0.1 H
1
Phenyl 3 5
CH3O H
F- -0.3 0.0 -0.2 H
Fluoro
Cl- 0.0 0.0 -0.1 H 6
Chloro 2H CH2
H 4 H
Br- 0.2 -0.1 0.0
Bromo 3 7
I- 0.4 -0.2 0.9
Iodo 1. (CH3) = 0.9 + 2.8 = 3.7
HO- -0.6 -0.1 -0.5 actual = 3.8
Hydroxy
RO- -0.5 -0.1 -0.4 2. (2) = 7.3 + (-0.5) ortho + (-0.1) para = 6.7
Alkoxy actual = 6.8
RCO 2- -0.3 0.0 -0.1
O-ester 3. (3) = 7.3 + (-0.2) ortho + (-0.4) para = 6.7
(H)2/R2N- -0.8 -0.2 -0.7 actual = 7.1
N-amino
RCONH- 0.1 -0.1 -0.3 4. (CH 2) = 1.2 + (0.8) + (1.4) = 3.4
N-amide actual = 3.3
O 2N- 1.0 0.3 0.4
Nitro 5. (5) = 5.2 + (0.7) gem = 5.9
RS- -0.1 -0.1 -0.2 actual = 5.9
thiol/sulfide
OHC- 0.6 0.2 0.3 6. (6) = 5.2 + (-0.2) trans = 5.0
Aldehyde
ROC- 0.6 0.1 0.2 actual = 5.1
Ketone
HO2C- 0.9 0.2 0.3 7. (7) = 5.2 + (-0.2) cis = 5.0
C-acid actual = 5.1
RO2C- 0.7 0.1 0.2
C-ester
H 2NOC- 0.6 0.1 0.2
C-amide
NC- 0.4 0.2 0.3
Nitrile
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 9
Real Examples of Combination Effects on Chemical Shifts
bond anisotropy 0.8 shielded bond example too
(CH 2)
0.8, shielded
H shielding cone
from bond
CH2
2.6
H deshielded H
7.2 1.5
electronegativity and bond 10-12 15, hydrogen
H bonded enol
O O H O O O
C H 9.5 C C C C
H 3C C CH3
O H O hydrogen
bonding H
electronegative substituent and distance from protons
O CH2CH 2CH2CH2CH 3 CH3 Cl CH3CH 2 Cl CH3CH2CH 2 Cl CH3CH 2CH 2CH2 Cl CH3CH 2 R
3.6 1.5 1.3 1.3 0.9 3.0 1.3 1.0 0.9 0.9
multiple substituents
CH4 CH 3Cl CH2Cl2 CHCl3 CCl4
0.2 = 2.8 3.0 = 2.3 5.3 = 1.9 7.2 =? ? (oops)
substituents at methyl (CH3), methylene (CH2) and methine (CH) O O O
CH3Cl CH3CH 2Cl (CH 3)2CHCl H 3C C Ph H 3CH2C C Ph (H3C)2CH C Ph
3.0 3.5 4.1 2.6 3.0 3.5
alkene substituent resonance and inductive effects
3.7 O
0.9 1.4 2.0 3.8 O 4.2 4.9
H H CH CH CH H 5.0 CH O C 6.4 C O H
3 2 2 H H 2C O H
C C 3
C C H 3C C C H 3C C C
C C 2.1
H H H H 1.3 H H H H
H H
5.3 5.8 4.9 6.1 5.8 6.5 4.0 7.3 4.6
aromatic resonance and inductive effects
H H 7.3 6.6 7.1 8.2 7.5
H H H H H H H H
6.7 O 7.7 O
H H
H 2N H H 2N H N H N H
O O
H H
bond anisotropy H H H H H H H H
produces deshielding Extra electron density via resonance produces Withdrawal of electron density via resonance
effect on aromatic shielding effect on aromatic protons, especially produces deshielding effect on aromatic protons,
protons. at ortho/para positions. especially at ortho/para positions.
sp C-H RO H alcohol H = 1-5 R2N H amine H = 1-5 enol H = 10-17 H O
H C C H 2.4 ArO H C C
phenol H = 4-10 O O
R C C H 1.9 RS H thiol H = 1-2.5 R C amide H = 5-8
R C
Ar C C H 3.0 ArO H aromatic thiol H = 3-4 NH2 O H acid H = 10-13
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 10
1. One nearest neighbor proton
increasing
one neighbor
H1 proton = Ha increasing E (, Bo)
Ha
C C the ratio of these
observed
perturbation(s) by two populations
proton neighbor proton(s) is about 50/50 (or 1:1)
Eto flip proton H1
E2 (observed)
Bo E1 (observed)
J1a
1 1
Protons in this environment have a small cancellation H1 H1 Protons in this environment have a small
of the external magnetic field, Bo, and produce a additional increment added to the external
smaller energy transition by that tiny amount. C C C C magnetic field, Bo, and produce a higher
energy transition by that tiny amount.
J = coupling constant N + 1 rule (N = # neighbors)
small difference in
energy due to differing J (Hz)
# peaks = N + 1 = 1 + 1 = 2 peaks
neighbor's spin (in Hz)
(ppm)
2. Two nearest neighbor protons (both on same carbon or one each on separate carbons)
the ratio of these
two neighbor four populations
H1
Ha protons is about 1:2:1
C C Hb H1
observed
proton Eto flip proton E1
E2 J1a
E3
Bo J1b J1b
two equal energy 1 2 1
H1 two neighbor protons are like
populations here
two small magnets that can be
C C arranged four possible ways
N + 1 rule (N = # neighbors)
(similar to flipping a coin twice)
J (Hz) J (Hz) # peaks = N + 1 = 2 + 1 = 3 peaks
(ppm)
3. Three nearest neighbor protons (on same carbon, or two on one and one on another, or one each on separate carbons)
three neighbor the ratio of these
H1
Ha protons eight populations
is about 1:3:3:1
C C Hb
observed H1
proton Hc
E1
Eto flip proton E2 J1a
E3
E4
Bo
J1b J1b
H1 three neighbor protons are like J1c J1c J1c
three small magnets that can be three equal energy 1 3 3 1
C C arranged eight possible ways populations at each
(similar to flipping a coin thrice) of middle transitions
N + 1 rule (N = # neighbors)
J (Hz) J (Hz) J (Hz)
# peaks = N + 1 = 3 + 1 = 4 peaks
(ppm)
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 11
Splitting patterns when the N+1 rule works (common, but not always true)
= group without any coupled proton(s)
N=0 N=1 N=2 N=3 H
H H H H H H H
H2 C CH2
C C C C C C CH C CH3 CH C CH
CH
CH CH
s, J=none d, J=7 t, J=7
I=1H q, J=7
I=1H I=1H I=1H
N=0 N=1 N=2
N=3
= calc or exp = calc or exp = calc or exp = calc or exp
N=4 N=5
H H
H H H H
CH C CH3 CH C CH2
C CH3 C CH2 CH C CH2 C CH3
CH CH2
CH CH2 CH CH2
qnt, J=7 sex, J=7
I=1H I=1H
N=4 N=5
= calc or exp = calc or exp
N=6 N=7
H H H
H H
C CH3 CH C CH3 H2C C CH2
CH C CH3 H2 C C CH3
CH3 H2 C CH2
CH3 CH2
oct, J=7
sep, J=7 I=1H
I=1H N=7
N=6
= calc or exp = calc or exp
N=8 H Pascal's triangle = coefficients of variable terms in binomial expansion (x + y)n, n = integer
Multiplets when the N + 1 rule works (all J values are equal).
H2C C CH3
1 peak = 100%
CH3 s = singlet 1 1 peak = 50%
d = doublet 1 1 1 peak = 25%
non, J=7
I=1H t = triplet 1 2 1 1 peak = 12% relative sizes of
N=8 q = quartet 1 3 3 1 peaks in multiplets
1 peak = 6%
qnt = quintet (% edge peak shown)
1 4 6 4 1 1 peak = 3%
sex = sextet 1 5 10 10 5 1 1 peak = 1.5%
= calc or exp sep = septet 1 6 15 20 15 6 1
o = octet 1 peak = 0.8%
1 7 21 35 35 21 7 1
Combinations or these are possible.
dd = doublet of doublets; ddd = doublet of doublet of doublets; dddd = doublet of doublet of doublet of doublets; dt = doublet of triplets
td = triplet of doublets; etc.
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 12
Typical Coupling Constants
Range Typical Range Typical
Ha Ha C Hb 0-3 Hz 1 Hz
C 0-30 Hz 14 Hz
C C
Hb
geminal protons - can have different chemical shifts cis / allylic coupling,
and split one another if they are diastereotopic notice through 4 bonds
Range Typical Range Typical
Ha Hb Ha
6-8 Hz 7 Hz C C 0-3 Hz 1 Hz
C C
C Hb
vicinal protons are on adjacent atoms, when freely trans / allylic coupling,
rotating coupling averages out to about 7 Hz notice through 4 bonds
Range Typical Range Typical
= dihedral C C
Ha Hb
angle
H 0-12 Hz 7 Hz C C 9-13 Hz 10 Hz
C C
Ha Hb
depends on dihedral
H angle, see plot of sp2 vicinal coupling
Karplus equation (different bonds)
Range Typical Range Typical
Ha Hb Ha Hb
0-1 Hz 0 Hz 1-3 Hz 2 Hz
C C C C C
O
protons rarely couple through 4 chemical bonds sp3 vicinal aldehyde coupling
unless in a special, rigid shapes (i.e. W coupling)
Range Typical Range Typical
Ha 0-3 Hz 2 Hz Ha Hb
5-8 Hz 6 Hz
C C C C
Hb C O
2
sp2 geminal coupling sp vicinal aldehyde coupling
Range Typical Range Typical
Ha Hb
5-11 Hz 10 Hz Ha
C C 2-3 Hz 2 Hz
C C C Hb
sp2 cis (acylic) coupling (always sp / propargylic coupling
smaller than the trans isomer) notice through 4 bonds
Range Typical Range Typical
Ha
C C Ha Hb
11-19 Hz 17 Hz 2-3 Hz 3 Hz
Hb C C C C
sp2 trans coupling (always bis-propargylic coupling
larger than the cis isomer) notice through 5 bonds
Range Typical ortho, meta and H Range Typical
Ha para coupling to
H ortho
C C 4-10 Hz 7 Hz this proton ortho 6-10 Hz 9 Hz
C Hb meta 2-3 Hz 2 Hz
H meta para 0-1 Hz 0 Hz
sp2 / sp3 vicinal coupling Hpara
When J values are less than 1 Hz, it is often difficult to resolve them and a peak may merely appear wider and shorter.
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 13
Similar chemical shift information presented in a different format. Remember, proton decoupled
carbons appear as singlets. When carbons are coupled to their hydrogens, carbons follow the N+1 rule.
Methyls = q, methylenes = t, methines = d, and carbons without hydrogen appear as singlets = s.
DEPT provides the same information. Carbon chemical shifts are spread out over a larger
range than proton chemical shifts (they are more dispersed), so it is less likely that two different carbon
shifts will fall on top of one another. The relative positions of various types of proton and
carbon shifts have many parallel trends (shielded protons tend to be on shielded carbons, etc.)
CH2 CH C
Simple alkane CH3
carbons
d 0 - 30 ppm d 20 - 40 ppm d 30 - 50 ppm d 30 - 60 ppm
(q) (t) (d) (s)
CH3 O CH2 O CH O
sp3 carbon C O
next to oxygen
d 50 - 60 ppm d 55 - 80 ppm d 60 - 80 ppm d 70 - 90 ppm
(q) (t) (d) (s)
CH2 N CH N
sp3 carbon CH3 N C N
next to nitrogen d 35 - 55 ppm
d 10 - 50 ppm
d 50 - 70 ppm d 50 - 70 ppm
(q) (t) (d) (s)
CH2 X CH X
sp3 carbon next to C X
bromine or chlorine
(X = Cl, Br) d 25 - 50 ppm d d 60 - 80 ppm
60 - 80 ppm
(t) (d) (s)
sp carbon (alkynes) C C sp carbon (nitriles) C N
70 - 90 ppm 110 - 125 ppm
sp2 carbon (alkenes
and aromatics) C H C X C X
C H
100 - 140 ppm 140 - 160+ ppm
2
simple sp2 carbon sp carbon attached to an electronegative atom
(X = oxygen, nitrogen, halogen) or C carbon
resonance donation moves lower,
conjugated with a carbonyl group
resonance withdrawal moves higher
O O O
C C C
X H R
160 - 180 ppm 180 - 210 ppm 180 - 220 ppm
carboxyl carbons aldehyde carbons, lower ketone carbons, lower
(acids, esters, amides) values when conjugated values when conjugated
(s) (d) (s)
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 14
Calculations of alkane 13C chemical shifts not listed above.
sp3 Carbon Chemical Shift Calculations
Calculations for sp3 carbon 13C chemical shifts of functionalized carbon skeletons can be performed starting
from the actual shifts found in the corresponding alkane skeleton, and introducing corrections factors based on the
functionality present in the molecule. This assumes that the alkane 13C shifts are available, which is why several
examples are provided below.
Examples of Cn alkanes as possible starting points for calculation 13C shifts in ppm.
Steric Corrections for sp3 carbon chemical shift calculations Approximate 13C shift calculation from scratch.
The attached C carbons are:
The calculated C = -(2) + 9x(# + #) - 2x(# ) + steric corrections
carbon atom is:
primary secondary tertiary quaternary
C1 = -2 + 9(1+3) - 2(2) + (-3) = 29 (actual = 28.3)
primary 0 0 -1.1 -3.4
C2 = -2 + 9(4+2) - 2(2) + [3x(-1.5)+(-15.0)] = 28 (actual = 34.0)
secondary 0 0 -2.5 -7.5 2 4 C3 = -2 + 9(3+5) - 0(2) + [(-9.5)+(-15.0)] = 45 (actual = 47.9)
3
0 -3.7 -9.5 -15.0 1 5
tertiary C4 = -2 + 9(3+2) - 3(2) + (-9.5) = 27 (actual = 27.2)
-1.5 -8.4 6
-15.0 -25.0 C5 = -2 + 9(1+2) - 2(2) + (-1) = 20 (actual = 19.5)
quaternary
C6 = -2 + 9(1+2) - 5(2) + (-1) = 14 (actual = 8.5)
13
C shifts for various carbon alkane skeletons - useful starting points for calculating sp3 carbon chemical shifts
22.3
C2 C3 C4 C5
16.3 25.4 22.6 32.0
25.0 30.2 11.8 29.7
CH4
25.0
-2.3 5.9 15.8 14.1 34.4 31.9 32.0
13.8
C6 29.0
22.7 11.5
22.9
19.5
27.9 20.6 8.9
30.4
14.1 32.9 18.8 33.9
36.3
41.5 14.4 29.3 36.5
19.2 29.8 14.7 15.0
48.3
C7 22.7 11.4 20.3
22.9 29.3 12.0
29.9 14.1 31.8
33.4 17.5 18.1
34.4 40.6
14.1 32.1 28.1 29.7 39.2 14.5 26.9
39.0 23.1
22.9 48.9 27.0
27.0
11.0 37.2 17.9
25.6 9.1
38.0
35.3 42.3
25.2
36.4
C8
14.5 39.6
22.9 29.5 22.7 29.6 36.5 19.3
27.2 22.8 34.6 10.9 40.3 20.1
20.3 32.4
14.1 32.1 28.1 11.5 29.7 14.1
39.2 32.3 14.1 25.6 35.4 14.6
19.7
C9 C10
22.8 29.6 22.9 29.6
14.1 32.1 29.8 14.2 32.2 29.9
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 15
R
X
X
X is attached to a terminal carbon atom (ppm) X is attached to an internal carbon atom (ppm)
Substituent = X C correction C correction C correction C correction C correction C correction
CH3 9 9 -2 6 8 -2
CH2CH3 18 7 -2 9 6 -2
CH(CH3)2 26 4 -2 14 3 -2
C(CH3)3 32 2 -2 20 1 -2
C
H
CH2 20 6 -1 15 5 -1
C CH 5 5 -4 2 6 -4
23 9 -2 17 7 -2
X is attached to a terminal carbon atom (ppm) X is attached to an internal carbon atom (ppm)
Substituent = X C correction C correction C correction C correction C correction C correction
OH 48 10 -6 44 7 -4
OR 60 7 -6 57 5 -6
O
51 6 -6 5 -6
O C 49
NH2 28 10 -5 24 8 -5
NH(CH3) 38 8 -5 32 5 -4
N(CH3)2
45 5 -5 37 3 -4
O
H
N C 26 7 -5 5 -5
21
R
NO2 62 5 -5 58 2 -5
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 16
X is attached to a terminal carbon atom (ppm) X is attached to an internal carbon atom (ppm)
Substituent = X C correction C correction C correction C correction C correction C correction
F 70 8 -7 67 5 -7
Cl 31 10 -5 36 8 -5
Br 20 10 -4 28 10 -4
I -7 11 -2 7 11 -2
O
C 30 0 -3 24 -1 -3
H
O
C 31 1 -3 26 0 -3
CH3
O
C 22 2 -3 18 1 -3
OH
X is attached to a terminal carbon atom (ppm) X is attached to an internal carbon atom (ppm)
Substituent = X C correction C correction C correction C correction C correction C correction
O
C 20 2 -3 16 2 -3
OCH3
O
C 25 3 -3 19 2 -3
NH2
C N 3 3 -3 3 3 -3
O
C 33 2 -3 30 2 -3
Cl
SH 11 10 -3 12 8 -3
SR 22 8 -3 20 6 -3
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 17
Additional starting point for calculating 13C chemical shifts (ppm) of substituted benzene rings (just a few possibilities)
Substituent Starting points for other common ring
systems. (ppm). No correction terms
128 ppm starting point for 1 included for substituents.
benzene carbon Use correction term for carbon
atom in relative position to the 126
4 2 substituent. Start with 128 ppm. 128
3 134
Substituent Z1 Z2 Z3 Z4 Substituent Z1 Z2 Z3 Z4
-H 0 0 0 0 -OH 29 -13 1 -7
-CH3 9 1 0 -3 -OCH3 34 -14 1 -8
-CH2CH3 12 -1 0 -3 -OC6H5 28 -11 0 -7 naphathalene
-CH2CH2CH3 10 0 0 -3 -NH2 18 -13 1 -10
136
-CH2CH2CH2CH3 11 0 0 -3 -NHCOCH3 10 -8 0 -4
-CH(CH3)2 20 -2 0 -3 -NHOH 22 -13 -2 -5
-C(CH3)3 19 -3 0 -3 -NHNH2 23 -16 1 -10 124
pyridine
-CH2F 8 -1 0 0 -N=N-R 22 -6 0 -3
-CH2Cl 9 0 0 0 -NO 37 -8 1 7 150
-CH2Br 9 1 0 0 -NO2 20 -5 1 6 N
-CH2I 11 -1 0 -1 -SH 4 1 0 -3 108
-CH2OH 12 -1 0 -1 -SCH3 10 -2 0 -4
-CH2NH2 15 -1 0 -2 -S(O)CH3 18 -5 1 2 pyrrole
-CH2NO2 2 2 1 1 -SO2CH3 12 -1 1 5 118
-CH2CN 2 0 -1 -1 -SO2Cl 16 -2 1 7 N
H
-CH2SH 12 -1 0 -2 -CN -16 3 1 4
-CH2CHO 7 1 0 -1 -CHO 8 1 0 6 110
-CH2COCH3 6 1 0 -2 -COCH3 9 0 0 4 furan
-CH2CO2H 6 1 0 -1 -CO2H 2 2 0 5
-CH2=CH2 13 -3 0 -1 -CO2CH3 2 1 0 4 143
O
-CCH -6 4 0 0 -CONH2 5 -1 0 3
-C6H5 8 -1 0 -1 -COCl 11 0 0 -3 126
-F 34 -13 2 -4 -Li -43 -13 2 3 thiophene
-Cl 5 0 1 2 -MgBr -36 -11 3 4
-Br -5 3 2 -1 125
-I -31 9 2 -1 S
Additional starting point for calculating 13C chemical shifts (ppm) of substituted alkenes (just a few possibilities)
123 ppm starting point for alkene carbon Z
C C
' ' ' 2 1 Effect of substituents on alkene 13C shifts (ppm)
C C C C C C C C
Substituent Z1 Z2 Substituent Z1 Z2
-H 0 0 -F 24 -34
C = 123 ppm + Zi -CH3 13 -7 -Cl 3 -6
123 + correction factors -CH2CH3 17 -10 -Br -9 -1
-CH2CH2CH3 16 -9 -I -38 7
increments for directly attached carbon atoms -OCH3 29 -39
-CH(CH3)2 23 -12
= 11 ' = -8 -C(CH3)3 26 -15 -O2CCH3 18 -27
=5 ' = -2 -CH2Cl 10 -6 -N(CH3 )2 28 -32
= -2 '= 2 -NO2 22 -1
-CH2Br 11 -5
-CH2I 14 -4 -CN -15 14
steric corrections -SCH2CH3 9 -13
for each pair of cis-, ' substituents -1 -CH2OH 14 -8
for each pair of geminal-, substituents -5 -CH=CH2 14 -7 -CHO 15 14
for each pair of geminal-, 'substituents 3 -CCH -6 6 -COCH3 14 5
if one or more sutstituents are present 2 -C6H5 12 -11 -CO2H 5 10
-COCl 8 14
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 18
Common fragmentation patterns in mass spectroscopy
1. Branch next to a bond
R
C C C C C C C C C
radical cation Pi electrons partially fill in loss of electrons at Characteristic carboncation stability also applies.
carbocation site via resonance. This is common
bond of an alkene fragmentation for alkenes and aromatics 3o R > 2o R > 1o R > CH3
or an aromatic
2. Branch next to an atom with a lone pair of electrons
R
X C X C X C
radical cation X lone pair electrons partially fill in loss of electrons at carbocation site via resonance. This
is a common fragmentation for any atom that has a lone pair of electrons (oxygen = alcohol,
ether, ester; nitrogen = amine, amide, sulfur = thiol or sulfide, etc.). Alcohols often lose water
(M-18) and primary amines can lose ammonia (M-17).
3. Branch next to a carbonyl (C=O) bondand possible subsequent loss of carbon monoxide, CO
R1 C O R1 C O R1
O
loss of
C
R1 R2
C O R2
radical cation R2 C O R2 C O
R1 or R2 can be lost from An oxygen lone pair partially fill in the loss of electrons at the subsequent loss of CO is possible
aldehydes, ketones, acids, carbocation site via resonance. This is a common fragmentation after fragmentation so not only can
esters, amides...etc. pattern for any carbonyl compound and can occur from either you see loss of an branch you can
side, though some are more common than others. see the mass of an branch.
4. McLafferty Rearrangement
Positive charge can be on
H H
O C C either fragment, which
O
typically have an even mass.
C C C C
R1 C R1 C = alpha position
= beta position
lost neutral
radical cation still a radical fragment = gama position
cation
This is another common fragmentation pattern for carbonyl compounds (and other pi systems as well: alkenes, aromatics,
alkynes, nitriles, etc.). If the pi bond has at least 3 additional nonhydrogen atoms attached and a hydrogen on the "gama"
atom, the branch can curve around to a comfortable 6 atom arrangement and the pi bond can pick up a hydrogen atom and
cut off a fragment between the C and C positions. The positive charge can be seen on either fragment and usually the
fragments have an even mass (unless there is an odd number of nitrogen atoms).
Knowing these few fragmentation patterns will allow you to make many useful predictions and
interpretations. Loss of small molecules, via elimination is common: H2O = 18, H2S = 34, CH3OH = 32,
C2H5OH = 46, NH3 = 17, CH3CO2H = 62, HF = 20, HCl = 36/38, HBr = 80/82, etc.
Z:\files\classes\spectroscopy\typical spectra charts.DOC
Spectroscopy Data Tables 19
A sampling of unusual and/or miscellaneous peaks that are commonly seen, (even when they don't make sense).
R R
CH3 = 15 R R
CH3CH2 = 29 C
C3H7 = 43 mass = 39 (R = H) H
C4H9 = 57 53 (R = CH3)
67 (R= CH2CH3) mass = 41 (R = H) mass = 65 (R = H) mass = 91 (R = H)
C5H11 = 71
also works for 55 (R = CH3) 79 (R = CH3) 105 (R = CH3)
C6H13 = 85
69 (R= CH2CH3) 93 (R= CH2CH3) 119 (R= CH2CH3)
R CH2
H
H2N C O R O C R
H
C
H2 mass = 29 (R = H)
mass = 44
43 (R = CH3)
mass = 42 (R = H)
H 57 (R= CH2CH3)
mass = 27 56 (R = CH3) 71 (R = C3H7)
mass = 77 RO C O
70 (R= CH2CH3) 105 (R = C6H5)
mass = 45 (R = H)
59 (R = CH3)
73 (R= CH2CH3)
Loss of small molecules via elimination reactions.
H2 O H2 S CH3OH C2H5OH NH3 CH3CO2H HF HCl HBr
80
mass = 18 34 32 46 17 62 20 36 82
38
McLafferty Possibilities
H
H
O R
O R2 McLafferty R2
Notice! HC
even masses C =
R1 R1
R CH2 CH2
R C variable mass,
H2 mass = 44 (R = H) mass = 28 (R = H)
58 (R = CH3) (can sometimes see 42 (R = CH3)
72 (R= CH2CH3) cation on this side too) 56 (R= CH2CH3)
86 (R = C3H7) 70 (R = C3H7)
Similar Patterns
H H
H
CH2 R2 H H
R2
C R2 N R2
R1
R1
R C R1 R1
H2 C
H2 C C
H2 H2
H H
H2C H H H
R2
H C R2 N
R2 R2
C R1 R
R CH2 C R1 C R1
R1
CH2 CH2
mass = 42 (R = H) mass = 40 (R = H) mass = 41 (R = H)
mass = 92 (R = H)
56 (R = CH3) 106 (R = CH3) 54 (R = CH3) 55 (R = CH3)
70 (R= CH2CH3) 120 (R= CH2CH3) 68 (R= CH2CH3) 69 (R= CH2CH3)
84 (R = C3H7) 134 (R = C3H7) 82 (R = C3H7) 83 (R = C3H7)
Z:\files\classes\spectroscopy\typical spectra charts.DOC
You might also like
- Spec Ir NMR Spectra TablesDocument15 pagesSpec Ir NMR Spectra TablesMah NovaesNo ratings yet
- Infrared (IR) Spectroscopy: Structure, Purity, and IdentityDocument16 pagesInfrared (IR) Spectroscopy: Structure, Purity, and IdentityDiana KowsariNo ratings yet
- IR ProcedureDocument5 pagesIR ProcedurePuvaneswary LoganathanNo ratings yet
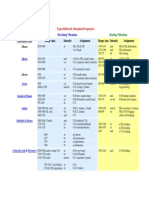
- Functional Class Range (NM) Intensity Assignment Range (NM) Intensity AssignmentDocument6 pagesFunctional Class Range (NM) Intensity Assignment Range (NM) Intensity AssignmentdubstepoNo ratings yet
- IR ProcedureDocument5 pagesIR ProcedureMuhammad FauziNo ratings yet
- Infrared Spectroscopy: Conformational IsomersDocument7 pagesInfrared Spectroscopy: Conformational IsomersRiyan NazarudinNo ratings yet
- Spektro IRDocument64 pagesSpektro IRAnonymous NSK4nvH4ufNo ratings yet
- 2230L 08 IR Spectra InterpretationDocument11 pages2230L 08 IR Spectra Interpretationvennilaj23No ratings yet
- ASS Instrumental OrganicDocument17 pagesASS Instrumental OrganicMohamed SakrNo ratings yet
- Infrared Spectroscopy: Concepts and TheoriesDocument55 pagesInfrared Spectroscopy: Concepts and Theoriesdead_knightNo ratings yet
- Lecture 4 IR Spectrum AnalysisDocument43 pagesLecture 4 IR Spectrum AnalysiskhadijahhannahNo ratings yet
- IR-freq CO BondDocument3 pagesIR-freq CO BondRD's AcademyNo ratings yet
- Raw Material Analysis-IRDocument58 pagesRaw Material Analysis-IRDilla Wulan NingrumNo ratings yet
- 08 - Infrared Spectroscopy ManualDocument5 pages08 - Infrared Spectroscopy ManualShubham BobadeNo ratings yet
- Infrared Correlations: Functional Group Band Position (CM) AppearanceDocument2 pagesInfrared Correlations: Functional Group Band Position (CM) AppearanceAmritansh RanjanNo ratings yet
- Key HW 3 Part II SpecDocument16 pagesKey HW 3 Part II SpecTha KantanaNo ratings yet
- PHR410 Chapter 2Document36 pagesPHR410 Chapter 2pulock.paulNo ratings yet
- Ir Func GroupDocument52 pagesIr Func GroupEry NourikaNo ratings yet
- 6-IR Spectroscopy of Alkane, Alkene and Carbonyl CompoundsDocument8 pages6-IR Spectroscopy of Alkane, Alkene and Carbonyl Compoundsbloodhound13042005No ratings yet
- Spec Ir NMR Spectra Tables PDFDocument15 pagesSpec Ir NMR Spectra Tables PDFYuppie RajNo ratings yet
- Introduction To Interpretation of Infrared SpectraDocument3 pagesIntroduction To Interpretation of Infrared SpectraBenni WewokNo ratings yet
- IR SPECTROSCOPY Notes FullDocument5 pagesIR SPECTROSCOPY Notes FullKartik KuteNo ratings yet
- Spectroscopy Infrared SpectraDocument51 pagesSpectroscopy Infrared Spectrathanasa08No ratings yet
- FTIR TablesDocument1 pageFTIR TablesvandykavidurgaNo ratings yet
- IR SpectrosDocument33 pagesIR SpectrosKikiMariaNo ratings yet
- IR Spectra AnalysisDocument37 pagesIR Spectra AnalysisdevoydouglasNo ratings yet
- Introduction To Interpretation of Infrared SpectraDocument3 pagesIntroduction To Interpretation of Infrared Spectrachinnirao100% (4)
- Topic 9 NotesDocument9 pagesTopic 9 NotesRitik YadavNo ratings yet
- IRSpectrum AnalysisDocument2 pagesIRSpectrum AnalysisDavid S. FrohnapfelNo ratings yet
- Common I R Absorption SDocument1 pageCommon I R Absorption SVisakha SureshNo ratings yet
- IR Spectroscopy TutorialDocument36 pagesIR Spectroscopy TutorialreddygrNo ratings yet
- Functional Groups Functional Groups: Functional Group G PDocument52 pagesFunctional Groups Functional Groups: Functional Group G PZenonissya Galwan BataraNo ratings yet
- Interpretation of Spectra of Different CompoundsDocument15 pagesInterpretation of Spectra of Different Compoundsmariam nawabNo ratings yet
- Spec IR Table For Common Chemical SymbolsDocument4 pagesSpec IR Table For Common Chemical SymbolsYoussef LatashNo ratings yet
- Printable Acrobat PDF File: Table of Characteristic IR AbsorptionsDocument3 pagesPrintable Acrobat PDF File: Table of Characteristic IR AbsorptionsImam Hadillah MuhfiNo ratings yet
- Table - 1: Characteristic Infrared Absorptions of Functional GroupsDocument1 pageTable - 1: Characteristic Infrared Absorptions of Functional GroupsAJIT CHAUDHARINo ratings yet
- CHMBD 449 - Organic Spectral: AnalysisDocument40 pagesCHMBD 449 - Organic Spectral: AnalysisIleana ManciuleaNo ratings yet
- Infrared SpectrosDocument110 pagesInfrared SpectrosBHARTI GAURNo ratings yet
- Study Notes-IR SpectrosDocument24 pagesStudy Notes-IR SpectrosakshantratwanNo ratings yet
- Ir PDFDocument1 pageIr PDFBartłomiej LesiszNo ratings yet
- CHMBD 449 - Organic Spectral: AnalysisDocument43 pagesCHMBD 449 - Organic Spectral: AnalysisIleana ManciuleaNo ratings yet
- Infrared Tutorial 2Document71 pagesInfrared Tutorial 2Hammo Ez AldienNo ratings yet
- IR ChartDocument2 pagesIR ChartNadiaa SafirraNo ratings yet
- Solomons Organic Chemistry Module IR TableDocument1 pageSolomons Organic Chemistry Module IR TableBenni WewokNo ratings yet
- Functional Class Range (CM) Intensity Assignment Alkanes: AlkenesDocument1 pageFunctional Class Range (CM) Intensity Assignment Alkanes: AlkenesStoica AlexandruNo ratings yet
- Spektrometri IRDocument51 pagesSpektrometri IRClarion 642No ratings yet
- FullDocument10 pagesFullAbdul Wahab KhanNo ratings yet
- IR SpectrosDocument44 pagesIR SpectrosVansh YadavNo ratings yet
- Alcohol: Functional Group Type of Vibration Characteristic Absorptions (CM) IntensityDocument2 pagesAlcohol: Functional Group Type of Vibration Characteristic Absorptions (CM) IntensityMuhammad Fadhila Ragil YogaNo ratings yet
- Scanning Electron Microscopy (SEM) With Energy Dispersive Spectroscopy (EDS) AnalysisDocument5 pagesScanning Electron Microscopy (SEM) With Energy Dispersive Spectroscopy (EDS) AnalysisAjeeth KumarNo ratings yet
- Infrared Spectroscopy: IR Absorptions For Representative Functional GroupsDocument3 pagesInfrared Spectroscopy: IR Absorptions For Representative Functional GroupsSaleem BashaNo ratings yet
- Measures Molecular Vibrations of Characteristic Functional GroupsDocument4 pagesMeasures Molecular Vibrations of Characteristic Functional GroupsLejNo ratings yet
- Spectroscopy Infrared SpectraDocument51 pagesSpectroscopy Infrared SpectraAakshi JairathNo ratings yet
- Table of Characteristic IR AbsorptionsDocument4 pagesTable of Characteristic IR Absorptionsأمالي أريفينNo ratings yet
- Schaum's Easy Outline of Organic Chemistry, Second EditionFrom EverandSchaum's Easy Outline of Organic Chemistry, Second EditionRating: 3.5 out of 5 stars3.5/5 (2)
- Critical Evaluation of Equilibrium Constants Involving 8-Hydroxyquinoline and Its Metal Chelates: Critical Evaluation of Equilibrium Constants in Solution: Part B: Equilibrium Constants of Liquid-Liquid Distribution SystemsFrom EverandCritical Evaluation of Equilibrium Constants Involving 8-Hydroxyquinoline and Its Metal Chelates: Critical Evaluation of Equilibrium Constants in Solution: Part B: Equilibrium Constants of Liquid-Liquid Distribution SystemsNo ratings yet
- Fourth International Conference on Non-Aqueous Solutions: Vienna 1974From EverandFourth International Conference on Non-Aqueous Solutions: Vienna 1974V. GutmannNo ratings yet
- Audio IC Circuits Manual: Newnes Circuits Manual SeriesFrom EverandAudio IC Circuits Manual: Newnes Circuits Manual SeriesRating: 5 out of 5 stars5/5 (1)
- Vibrational Spectra of Organometallics: Theoretical and Experimental DataFrom EverandVibrational Spectra of Organometallics: Theoretical and Experimental DataNo ratings yet
- 103 NOT OUT Organic ChemistryDocument1 page103 NOT OUT Organic ChemistryJeevan KumarNo ratings yet
- Beggs - Brill Method PDFDocument12 pagesBeggs - Brill Method PDFTiago Mendes TavaresNo ratings yet
- Halogen TrapDocument9 pagesHalogen TrapSolano Mena100% (1)
- Density Estimation For Fatty Acids and Vegetable OilsDocument6 pagesDensity Estimation For Fatty Acids and Vegetable OilscymyNo ratings yet
- Module 1 - Organic ChemistryDocument12 pagesModule 1 - Organic ChemistrySelena MoonNo ratings yet
- GRP Brochure AmiantitDocument6 pagesGRP Brochure Amiantitbhathiya8No ratings yet
- Contents DrugDocument6 pagesContents Drugwestcoastaromatics0% (1)
- Edibon Fixed and Fluidized Bed ManualDocument40 pagesEdibon Fixed and Fluidized Bed ManualArianne BatallonesNo ratings yet
- Germanium-Based Integrated Photonics From Near-To Mid-Infrared ApplicationsDocument13 pagesGermanium-Based Integrated Photonics From Near-To Mid-Infrared ApplicationsMiguel Serrano RomeroNo ratings yet
- Ai TS 1 - Class XII - SET ADocument18 pagesAi TS 1 - Class XII - SET ASantryuNo ratings yet
- Apéndice de FoglerDocument18 pagesApéndice de FoglerBRIGITH STHEFANIA BENITES GARCIANo ratings yet
- Sodium Hydroxide BookletDocument24 pagesSodium Hydroxide BookletbbmokshNo ratings yet
- J.J.E. Herrera Velázquez Et Al - Instability Suppression by Sheared Flow in Dense Z-Pinch DevicesDocument8 pagesJ.J.E. Herrera Velázquez Et Al - Instability Suppression by Sheared Flow in Dense Z-Pinch DevicesImsaa4No ratings yet
- Dupont Zytel Htn51g35hsl Nc01Document6 pagesDupont Zytel Htn51g35hsl Nc01josebernal_mzaNo ratings yet
- Cheat Sheet Modelling 1718Document2 pagesCheat Sheet Modelling 1718Siti MaisarahNo ratings yet
- Experiment No 9 mm2 PDFDocument9 pagesExperiment No 9 mm2 PDFMuhammad Zeeshaan JavedNo ratings yet
- Phytoplankton Culture For Aquaculture Feed: S R A CDocument16 pagesPhytoplankton Culture For Aquaculture Feed: S R A CFen TZNo ratings yet
- Heat Transfer Question PaperDocument2 pagesHeat Transfer Question Paperbharathkumar0310No ratings yet
- Atomic Structures Formulas and NamesDocument4 pagesAtomic Structures Formulas and NamesKyla Mari ValduezaNo ratings yet
- Overview This Chapter Presents The Principles of Diffusion and ReactionDocument5 pagesOverview This Chapter Presents The Principles of Diffusion and ReactionCurieNo ratings yet
- Drying Technology - An Overview - For ME5202Document134 pagesDrying Technology - An Overview - For ME5202mahe_sce4702No ratings yet
- PaperDocument35 pagesPaperRashmi SharmaNo ratings yet
- Lesson 3 Properties of Fresh Concrete and Factors Affecting PropertiesDocument6 pagesLesson 3 Properties of Fresh Concrete and Factors Affecting PropertiesajaytrixNo ratings yet
- Southern Blotting TechniqueDocument5 pagesSouthern Blotting TechniqueVĩnh Nguyễn VănNo ratings yet
- Vitamin C: Science Topics Process Skills VocabularyDocument7 pagesVitamin C: Science Topics Process Skills VocabularyJin Xiai TianNo ratings yet
- MSDS H2oDocument5 pagesMSDS H2oWilanda Tama50% (2)
- Optical SourceDocument46 pagesOptical Sourcemanishsoni30100% (1)
- Problema 12-10 TreybalDocument1 pageProblema 12-10 TreybalMiguel Angel Lugo CarvajalNo ratings yet
- Oxidation NumberDocument21 pagesOxidation NumberChristian LopezNo ratings yet
- Animal Manure - 2020 - Waldrip - Organomineral Fertilizers and Their Application To Field CropsDocument15 pagesAnimal Manure - 2020 - Waldrip - Organomineral Fertilizers and Their Application To Field CropsEduardo GuimaraesNo ratings yet