Professional Documents
Culture Documents
Protein-Profile HMMS: Matching
Uploaded by
juanpapoOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Protein-Profile HMMS: Matching
Uploaded by
juanpapoCopyright:
Available Formats
Protein- Profile HMMs
As we have seen earlier, protein structural similarities make it possible to create a statistical model of
a protein family which is called a profile. The idea is, given a single amino acid target sequence of
unknown structure, we want to infer the structure of the resulting protein. The profile HMM is built
by analyzing the distribution of amino-acids in a training set of related proteins. This HMM in a
natural way can model positional dependant gap penalties.
The basic topology of a profile HMM is shown above. Each position, or module, in the model has
Matching Deletion
Insertion
three states. A state shown as a rectangular box is a match state that models the distribution of letters
in the corresponding column of an alignment. A state shown by a diamond-shaped box models
insertions of random letters between two alignment positions, and a state shown by a circle models
a deletion, corresponding to a gap in an alignment. States of neighboring positions are connected,
as shown by lines. For each of these lines there is an associated `transition probability', which is the
probability of going from one state to the other.
The match state represents a consensus amino acid for this position in the protein family. The delete
state is a non-emitting state, and represents skipping this consensus position in the multiple
alignment. Finally, the insert state models the insertion of any number of residues after this consensus
position.
You might also like
- 1.1. An Example of A HMM For Protein Sequences: Output ProbDocument16 pages1.1. An Example of A HMM For Protein Sequences: Output ProbjuanpapoNo ratings yet
- Training ProblemDocument9 pagesTraining ProblemjuanpapoNo ratings yet
- Training ProblemDocument12 pagesTraining ProblemjuanpapoNo ratings yet
- Three Problems of Hidden Markov Models: 1) Scoring ProblemDocument11 pagesThree Problems of Hidden Markov Models: 1) Scoring ProblemjuanpapoNo ratings yet
- Pairwise Sequence AlignmentDocument12 pagesPairwise Sequence Alignmentvanigo1824No ratings yet
- Bio Informatics 005Document4 pagesBio Informatics 005Arooba ShujaNo ratings yet
- Homolgy ModelingDocument19 pagesHomolgy ModelingJainendra JainNo ratings yet
- Homology Modeling, Also Known As Comparative Modeling ofDocument19 pagesHomology Modeling, Also Known As Comparative Modeling ofManoharNo ratings yet
- Protein Structure Similarity: Mlesnick@stanford - EduDocument8 pagesProtein Structure Similarity: Mlesnick@stanford - EdualsreshtyNo ratings yet
- Methods For Applying Multiple Sequence AlignmentDocument17 pagesMethods For Applying Multiple Sequence AlignmentacezeeshanNo ratings yet
- On The Entropy of Protein Families: John P. Barton Arup K. Chakraborty Simona Cocco Hugo Jacquin Rémi MonassonDocument27 pagesOn The Entropy of Protein Families: John P. Barton Arup K. Chakraborty Simona Cocco Hugo Jacquin Rémi MonassonDr. K Lalitha GuruprasadNo ratings yet
- Protein ModellingDocument53 pagesProtein ModellingiluvgermieNo ratings yet
- Multiple Sequence Alignment MSADocument8 pagesMultiple Sequence Alignment MSAAnonymous nUiP53SNo ratings yet
- Comparison of The PAM and BLOSUM Amino Acid Substitution MatricesDocument4 pagesComparison of The PAM and BLOSUM Amino Acid Substitution Matricesmorteza hosseiniNo ratings yet
- Protein Contact Prediction From Amino Acid Co-Evolution Using Convolutional Networks For Graph-Valued ImagesDocument9 pagesProtein Contact Prediction From Amino Acid Co-Evolution Using Convolutional Networks For Graph-Valued ImagesThao LeNo ratings yet
- Summary Bioinformation TechnologyDocument15 pagesSummary Bioinformation TechnologytjNo ratings yet
- Lecture 3 and 4 LSM2241Document6 pagesLecture 3 and 4 LSM2241Koh Yen SinNo ratings yet
- H ' I P P A: MM S Nterpolation of Rotiens For Rofile NalysisDocument10 pagesH ' I P P A: MM S Nterpolation of Rotiens For Rofile NalysisijcseitNo ratings yet
- Bioinformatics IDocument39 pagesBioinformatics IJan Patrick PlatonNo ratings yet
- Bif401 Solved Final Papers 2017Document8 pagesBif401 Solved Final Papers 2017HRrehmanNo ratings yet
- Bookmark This PageDocument35 pagesBookmark This PageAndreea SpiridonNo ratings yet
- Torsion Angles and The Ramachandran PlotDocument2 pagesTorsion Angles and The Ramachandran PlotRitika PaulNo ratings yet
- CME 4425 Introduction To Bioinformatics Algorithms: Zerrin Işık Zerrin@cs - Deu.edu - TRDocument23 pagesCME 4425 Introduction To Bioinformatics Algorithms: Zerrin Işık Zerrin@cs - Deu.edu - TRMahmud KorkmazNo ratings yet
- DH DerivDocument7 pagesDH Derivأحمد دعبسNo ratings yet
- LinearlyDocument12 pagesLinearlywangxiangwen0201No ratings yet
- Chap.3 Protein Structure & Function: TopicsDocument30 pagesChap.3 Protein Structure & Function: TopicssimalihaNo ratings yet
- Local and Global Alignment ResearchpaperDocument12 pagesLocal and Global Alignment ResearchpaperSam GlassmanNo ratings yet
- Polymer StructureDocument4 pagesPolymer StructurePradeep ChaudhariNo ratings yet
- 1 - Sequence Alignment and VisualizationDocument68 pages1 - Sequence Alignment and VisualizationAlisha100% (1)
- Substitution MatrixDocument10 pagesSubstitution MatrixRashmi DhimanNo ratings yet
- Lab Report 1 BME 310Document7 pagesLab Report 1 BME 310Can MunganNo ratings yet
- 322 Refrensi 19Document4 pages322 Refrensi 19bisniskuyNo ratings yet
- Protein3dstructureprediction 161109084221Document30 pagesProtein3dstructureprediction 161109084221Jhanvi SNo ratings yet
- CATH, Bilogical Data Bases, Bioinformatics Data BaseDocument3 pagesCATH, Bilogical Data Bases, Bioinformatics Data BaseRajesh GuruNo ratings yet
- Structure of ProteinDocument72 pagesStructure of ProteinSudipta MandolNo ratings yet
- Iguico, Maria SofiaDocument4 pagesIguico, Maria SofiaDelvin Jan del RosarioNo ratings yet
- Lec06 MPH46 5103Document21 pagesLec06 MPH46 5103Sabith Hasan Khan JoyNo ratings yet
- Bioinformatics Notes - 17Bt54: Module - 4Document48 pagesBioinformatics Notes - 17Bt54: Module - 4Samuel PrinceNo ratings yet
- Alignment CorrectionDocument3 pagesAlignment CorrectionfaisalnadimNo ratings yet
- Using Profile HMM in MSADocument4 pagesUsing Profile HMM in MSAedifoeNo ratings yet
- 7: Source-Based Nomenclature For Copolymers (1985)Document16 pages7: Source-Based Nomenclature For Copolymers (1985)darshanNo ratings yet
- Biochemistry LN05-1Document14 pagesBiochemistry LN05-1Rahaf Al-muhtasebNo ratings yet
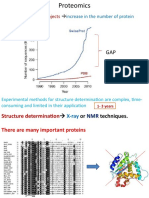
- Genome Sequencing Projects: Increase in The Number of Protein SequencesDocument27 pagesGenome Sequencing Projects: Increase in The Number of Protein SequencesBiju ThomasNo ratings yet
- Mustang PSFB FinalDocument49 pagesMustang PSFB FinalNiranjan BhuvanaratnamNo ratings yet
- On The Accuracy of Homology Modeling and Sequence AlignmentDocument10 pagesOn The Accuracy of Homology Modeling and Sequence AlignmentCRISTIAN GABRIEL ZAMBRANO VEGANo ratings yet
- Dot PlotDocument2 pagesDot PlotdarshowlkatNo ratings yet
- L5-Pairwise Seq AlignmentDocument69 pagesL5-Pairwise Seq AlignmentSwayamNo ratings yet
- Amino Acid Substitution Scores: 1 2 N 1 2 N N I 1 I IDocument3 pagesAmino Acid Substitution Scores: 1 2 N 1 2 N N I 1 I ISomnathNo ratings yet
- Protein Structure PredictionDocument52 pagesProtein Structure PredictionMudit MisraNo ratings yet
- Polymer StructureDocument4 pagesPolymer StructureVenkat Reddy YedullaNo ratings yet
- 2013 J Theor Biol 328 77-88Document12 pages2013 J Theor Biol 328 77-88mbrylinskiNo ratings yet
- Protein ModellingDocument15 pagesProtein ModellingNandini KotharkarNo ratings yet
- Statistical Measures To Quantify Similarity Between Molecular Dynamics Simulation TrajectoriesDocument17 pagesStatistical Measures To Quantify Similarity Between Molecular Dynamics Simulation TrajectoriesDiego GranadosNo ratings yet
- Protein Modelling: (Building 3D Models of Proteins)Document19 pagesProtein Modelling: (Building 3D Models of Proteins)rednriNo ratings yet
- Samudrala 1998aDocument22 pagesSamudrala 1998awangxiangwen0201No ratings yet
- Bioinformatics in PAM AND BLOSUMDocument17 pagesBioinformatics in PAM AND BLOSUMgladson100% (15)
- Identification of MOTIFS in Bioactive Peptides PrecursorsDocument14 pagesIdentification of MOTIFS in Bioactive Peptides PrecursorsijbbjournalNo ratings yet
- EOQ ModelsRDocument5 pagesEOQ ModelsRjuanpapoNo ratings yet
- Multiple Sequence AlignmentDocument16 pagesMultiple Sequence AlignmentjuanpapoNo ratings yet
- Future Work.Document20 pagesFuture Work.juanpapoNo ratings yet
- 1.1. Multiple Sequence AlignmentDocument16 pages1.1. Multiple Sequence AlignmentjuanpapoNo ratings yet
- Background: 1.1. DNA - Deoxyribonucleic AcidDocument19 pagesBackground: 1.1. DNA - Deoxyribonucleic AcidjuanpapoNo ratings yet
- Hidden Markov Model (HMM) ArchitectureDocument15 pagesHidden Markov Model (HMM) ArchitecturejuanpapoNo ratings yet
- Primary StructureDocument21 pagesPrimary StructurejuanpapoNo ratings yet
- Limitations of HMMS: P (X P (YDocument1 pageLimitations of HMMS: P (X P (YjuanpapoNo ratings yet
- P (X P (Y: 1. Open Areas For Research in Hmms in BiologyDocument1 pageP (X P (Y: 1. Open Areas For Research in Hmms in BiologyjuanpapoNo ratings yet
- The Central Dogma of Molecular Biology: CctgagccaactattgatgDocument18 pagesThe Central Dogma of Molecular Biology: CctgagccaactattgatgjuanpapoNo ratings yet
- RusobioloDocument21 pagesRusobiolojuanpapoNo ratings yet
- HMM ReportDocument27 pagesHMM ReportŽeljko ŽakulaNo ratings yet
- Biological Background: 1.1. DNA - Deoxyribonucleic AcidDocument19 pagesBiological Background: 1.1. DNA - Deoxyribonucleic AcidjuanpapoNo ratings yet
- Hidden Markov Model (HMM) ArchitectureDocument15 pagesHidden Markov Model (HMM) ArchitecturejuanpapoNo ratings yet
- Biology M PDFDocument13 pagesBiology M PDFjuanpapoNo ratings yet
- Activity Guide and Rubric - Act. 1 Recognition ForumDocument7 pagesActivity Guide and Rubric - Act. 1 Recognition ForumLina Mosquera GuzmanNo ratings yet