Professional Documents
Culture Documents
CRISPR Screen Workflows
CRISPR Screen Workflows
Uploaded by
scribdusername_mooOriginal Description:
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
CRISPR Screen Workflows
CRISPR Screen Workflows
Uploaded by
scribdusername_mooCopyright:
Available Formats
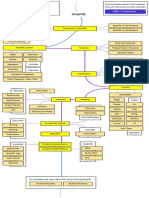
CRISPR Screen In Vitro Steps with IT Capability Needed
LentiViral sgRNA Establish Cas9+ Cell Meta Data
Generate Pool Create Arms IT Capability Example Data Output
Library LInes (MVP)
sgRNA Library
START # of guides
Perturbation Sequences Registers sgRNA Library
Perturbation Type with ID to track
sgRNA library with
Register Vector
Vector backbone
Vector Map
Source Registers Vector
Antibiotic resistance with ID to track
CRISPR Function: KO, KD etc
Cas9 (Yes/No)
pDNA
sgRNA Library ID Registers pDNA with
Vector ID ID to track
sgRNA Library
Barcode Barcoded tubes with
Inventory
Amount (volume) location
Location
Vector
Barcode Barcoded tubes with
Amount (volume) location
Location
pDNA
Barcode Barcoded tubes with
Amount (volume) location
START
Location
Parent Cell line
Cell line Name Registered Cell Line
Make Virus with ID to track
Register
Virus
Cell line ID
pDNA ID
Seeding density
Doubling time
Infectability of cell line
Virus batch ID
Method used to establish line
Registered Virus
Pool or Clone
with ID to track
Clone info
Details for establishing line
Antibiotic type
Antibiotic Concentration
Cas9 Assessment method
% Expression
Cells Ready
%Activity
Activity Data
Cell Line
Barcode Barcoded tubes with
Inventory
Amount (volume) location
Location
Virus Batch
Barcode Barcoded tubes with
Amount (volume) location
Location
Titer of Virus Data Scale up Cell Mix calculations (per rep)
Cell Line ID Minimium wells all reps 107
Media used .+4 wells for no infection control and pipetting 111
Cells to infect /rep 3.33E+08
Virus Batch
Media (mL) without virus 216.45
Polybrene Conc:
Assess Virus Total volume with virus (mL) 222
Instrument Capture Other Infection Details Vicell Polybrene (ug) 888
Protocol used Manual Entry
Titer Assessment Scale up Infection Mix (per rep) calculations
Volume virus Wells to infect (min +1) 108
Expected Titer Functional Volume Media+Cells+PB Mix (mL) 210.6
Expected Titer Flow by Volume Total virus vol (mL) 5.4
vol to add to 12 wells / well (mL) 2
Cell Line Name
START Source: Vendor / inhouse
CAS9 source Registered Cell Line
Register QC mycoplasma Test with ID to track
QC Cell line Identity
Verify Cas9 Expression
Grow & Passage Cells
(with or without Cas9)
Barcode
Barcoded tubes with
Inventory Amount
location
Location
Verify Cas9 expression
(if built in house)
Instrument Capture Labware: Flasks, 96 well FACS Data
Cells Ready? (If plate multiple cell lines, 3
wells/line with reference
cells
Cell Line Name
Source: Vendor / inhouse
CAS9 source Registered Cell Line
YES QC mycoplasma Test with ID to track
QC Cell line Identity
Verify Cas9 Expression
Cell Line A375
Infection Format 12 well high density Total volume/well (uL) 2000
Infection Batch Registered Infection Cells/Well 3.00E+06 Max Viral Input (uL) 50
Polybrene (ug/mL) 4 Total Resuspended Cell/well (uL) 1950
Register Cell line ID Batch with ID to Total vol/well (mL) 2
Virus Batch ID track
Infection Stage Wells 120 Polybrene (ug) 960
Cells 3.60E+08 Polybrene stock (ug/uL) 10
Total Cells Cell Master Mix (mL) 234 Polybrene (uL) 96
Total yield Polybrene (uL) 96
Instrument Capture Date of experiment
Time virus added Cell Mix (mL) Virus (mL) Media (mL)
& Virus + cells (Master mix) 218.4 5.6 0
Assay Capture Time virus Removed
ViCell NIC 1.95 0 0.05
Labware- Flasks or Dish
CS JL
Polybrene concentration Infect cells and spin at 930x g
9/23/2019 14:45 9/23/2019 14:45
30C for 2hr
Titered cell Move to incubator 9/23/2019 16:45 9/23/2019 16:45
Polybrene Pool cells and plate in 5 stacks 9/24/2019 11:00 9/24/2019 12:00
Labware 12 well Passage into puro 9/25/2019 10:30 9/25/2019 10:30
9/13/2019
Total Cells count in & out of each RFP+ (SSC-A v. PE-A) Cell Counts
(+) Puro (-) Puro
9/13/2019 9/16/2019 9/13/2019 9/16/2019
passage NIC 0.33% 0.80% 0.92% 1.1% NIC 0.00358 0.390 0.92%
400uL 0.0751 0.354 21.21%
Media volume ViCell 400uL 90.71% 89.59% 21.21% 20.1%
200uL 0.149 0.312 47.76%
200uL 66.12% 67.97% 47.76% 28.5%
Instrument Capture Puro (+)(-) Reference
100uL 40.48% 44.68% 8.66% 0.0%
Reference
100uL 0.0227 0.262 8.66%
Antibiotic Selection Date of experiment Virus (sgGFP)
50uL 23.30% 27.49% 8.91% 1.8%
Virus (sgGFP)
50uL 0.0262 0.294 8.91%
Stage & 25uL 12.17% 14.21% 7.78% 0.5% 25uL 0.0238 0.306 7.78%
Assay Capture Time selection started 12.5uL 5.92% 8.13% 2.29% 0.2% 12.5uL 0.00954 0.416 2.29%
6.25uL 3.56% 5.54% 5.22% 1.2% 6.25uL 0.0167 0.320 5.22%
Passage times 400uL 89.72% 90.09% 52.53% 49.8%
400uL 0.249 0.474 52.53%
Time selection end 200uL
100uL
71.03%
48.86%
73.88%
51.11%
50.00%
20.41%
28.7%
9.5%
200uL 0.139 0.278 50.00%
80K_hGwKO 100uL 0.0596 0.292 20.41%
Labware: 6 well plate or Flask/dish _p0
50uL 29.21% 36.15% 16.12% 2.0% 80K_hGwKO
0.0382 0.237
Cytoflex 25uL 17.20% 21.04% 11.55% 1.5% _p0
50uL 16.12%
12.5uL 7.90% 10.76% 5.44% 0.2% 25uL 0.0298 0.258 11.55%
6.25uL 3.93% 3.83% 2.59% 0.6% 12.5uL 0.0131 0.241 5.44%
NIC 0.8% 6.25uL 0.00835 0.322 2.59%
Register each Arm
Selection Registered arm with
Register Small molecule Conc. Used ID to track
Conditions
Create & Maintain Cell Pellets of each Arm
Arm 1 to many (4) Inventory Barcode Inventory Pellet
Amount
location
Screen background:
Purpose Total Reseed
Representation
Since
Doublings
Through
Day Day of Week Screen action Rep Conditions Conc Volume Cells reseeded Pre-Puro Post-Puro Post-puro
Optimization 0 Monday Infection CS
cells
3.20E+08
Vol
3.20E+08 1200 4000
Infection Puro
0
Generate Unique screen id 0.9
1.9
Tuesday
Wednesday
Pool
Split to Puro
CS
CS
3.80E+05
9.69E+05
1000
500
3.8E+08
4.85E+08
1000
500
3.80E+08
4.85E+08
1425
1817
4750
6056
0.25
0.60 0.35
4 Friday Passage on Puro CS 1.51E+06 510 7.7E+08 130.5 1.97E+08 2888 9626 1.27 1.02
Cell Line ID 7.1 Monday Start Co-Cultures CS 1.75E+06 502 8.79E+08 82.3 1.44E+08 3294 10981 3.42 3.18
Reference Virus ID 9 Wednesday End Co-culture CS
IFNy
No_IFNy
T-cells
E:T_0 1.23E+06 502 6.17E+08 0.00E+00 2315 7718 5.52 5.28 2.10
Reference Titer ID 9 Wednesday End Co-culture CS Yes_IFNy E:T_0 9.12E+05 501 4.57E+08 0 1713 5711 5.09 4.84 1.67
Instrument Capture 9 Wednesday End Co-culture CS No_IFNy E:T_2 2.43E+05 503 1.22E+08 0 458 1528 3.19 2.94 -0.24
Per Arm 9 Wednesday End Co-culture CS Yes_IFNy E:T_2 1.87E+05 505 9.44E+07 0 354 1180 2.81 2.57 -0.61
& ViCell
Morphology change
Assay Capture
Time tracked for each passage time IFNy T-cells
No_IFNy E:T_0
Rep
CS
IFNy T-cells
Yes_IFNy E:T_0
Rep
CS
IFNy
No_IFNy
T-cells
E:T_2
Rep
CS
IFNy T-cells
Yes_IFNy E:T_2
Doublings Doublings Doublings Doublings
Time track for pellet harvest or Screen end From
Through Puro Post-Puro Day
From Through
Post-Puro Day From Infection
Through
Post-Puro Day
From Through
Infection Infection Puro Puro Infection Puro
Total Cells 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0.247928 0 0 0.916667 0.247928 0 0 0.9166667 0.247927513 0 0 0.91666667 0.2479275 0
Total Media volume 0.598425 0.350497247
1.266974 1.019046378
0
0
1.916667
4
0.598425 0.350497
1.266974 1.019046
0
0
1.9166667
4
0.598424761 0.35049725
1.266973892 1.01904638
0
0
1.91666667
4
0.5984248 0.350497
1.2669739 1.019046
Viability 3.423418 3.175490308
5.52 5.28
0
2.10
7.083333
9
3.423418 3.17549
5.09 4.84
0
1.67
7.0833333
9
3.423417821 3.17549031
3.19 2.94
0
-0.24
7.08333333
9
3.4234178 3.17549
2.81 2.57
Day of Experiment
Labware: Flask /dish
Register DNA Pellet each arm Conc Post-Clean up Sample
Total Yield (ng) Representation (X) 11ug Aliquot
(ng/uL) Size
Cell Pellet ID Total Approx.
Cells of input Registered DNA with Sample
Representation
Nanodro
Qubit Volume
Nanodro
Qubit
Nanodro
Qubit
Nanodro
Qubit
By lowest
Register ID to track # p p p p quant
Preparation method used pre-clean up (mL)
1 1459.85 187 182 4 748000 728000 1416.667 1378.788 58.8 60.4 60.4
Modifications
2 1304.83 152.3 130 4.5 685350 585000 1298.011 1107.955 72.2 84.6 84.6
3 1360.23 192.1 146 3.6 691560 525600 1309.773 995.4545 57.3 75.3 75.3
DNA Pellet 4 1443.56 181.8 155 5 909000 775000 1721.591 1467.803 60.5 71.0 71.0
5 902.65 214.2 198 2 428400 396000 811.3636 750 51.4 55.6 55.6
DNA Pellet each arm 6 1654.26 184.5 154 4.5 830250 693000 1572.443 1312.5 59.6 71.4 71.4
Barcode Inventory Pellet
7 358.33 88.45 63 2.5 221125 157500 418.7973 298.2955 124.4 174.6 174.6
Inventory 8 1067.05 164.4 109 5 822000 545000 1556.818 1032.197 66.9 100.9 100.9
Amount
END location Sample gDNA
Rep T-cells IFNγ
# prep
1 CS No No 1,2 A+B
Data Capture each Pellet 2 JL No No 3,4 A+B
3 CS No Yes 5,6 A+B
DNA Pellet ID 4 JL No Yes 7,8 A+B
Assessment Method 5 CS Yes No 9 A+B
NanoDrop Concentration 6 JL Yes No 10,11 A+B
Instrument Capture Qubit Concentration Nanodrop 7 CS Yes Yes 12 A+B
8 JL Yes Yes 13,14 A+B
& Volume (manual)
Assay Capture Total yield
Total cells represented
Approximate library Qubit
representation
Labware: tube
CrispR Screen
Phase
INFy
Pick seq. guides LentiVirus
Sources
Vector map Other
(sgRNA 80K) sequence This is Human
Virus Optional step PBMC
Pick
Methodology Vector Cell Line Small
Vector Molecule
Data File Features Library And/OR
(txt)
START Activate TCells
Cell Line QC & CAS9 Compare retested
NO
Process
validation results vs original
Manufacture Assign Does viral
Pick Pick Vector Generate Infect cells Maintain
START
(Backbone) OR pDNA with vector Vectored
Lentivirus
Add Virus Titer Virus backbone have YES Validate Cas9 Cell line QC
with Library Selection Create ARMS
FEED ALL End Media
Create Pellets
Genomic DNA
Library & Library library Samples Genomics END
Cas9? ARMS Screen Removal Prep
NO
No YES
YES
Screening
Complete?
Selection
Complete?
Assay Types
In
DNA Functional Infection In
QC Selection Assay
Concentration Titer Screening
Media Viability Selection Method
Verify Cas9 Cell Cell Count Media Conc &
Growth Curve Cell Counts MycoPlasma Polybrene Infection Volume By Titer & Amount Cell Count
OR Expression Identity Conc
Total Cells Total Virus
Method Volume Viability Volume
Total Yield
NanoDrop Qubit
Provided by
Incucyte ViCell OR
manufacturer ViCell OR ViCell
Cas9
Volumetric or Combine cell, virus, &
Activity Fortess Cytoflex ViCell
Weighed on scale media to well (12 well
Instrument
GFP(NEW) Incucyte Fortess Cytoflex OR
plate)
Centrifuge
Remove & culture
NanoDrop
Qubit
Vector Features to track: Register Calculate & Graph
Register Data from ATCC Look up original QC
Antibiotic Resistance Virus Register in Excel or PRISM Record data Data from INDEXXX Record data
pDNA is source to data & compare
Function (KD, KO, activation) Virus in Excel or Charles River in Excel
compare results against new QC data Record data Inventory (-80)
Guide-only or All in one BioReg in Excel Record data Excel
Generation Associate information below in Excel
Vector map/features Prep method with registered cell line:
IT
Guides Volume generated % Survival
Concentration +/- selection by viral input
Selection used
Length of selection Record data
Infection method in Excel
SAMPLE ASSAY
SAPIO ARIA STUDYLOG DATA LAKE DARE/IJC
REQUEST APPS
Electrophoresis – Spectrophotmeter
Fluorometer –
QC DNA, RNA – measure DNA,
quantify DNA
samples RNA concentration
Upload list of Samples
using an Excel Template
TapeStation Nano-drop Qubit
Fluorescence
Video imaging
Pull in any general Sequencing activated cell
Select Samples from the Perform any automated system to analyze
sorting
Any pre ARIA “Create Study” in ARIA - Enter Notebook # & information about the Trigger an Assay Add the list of Samples Send Samples to SAPIO, cells
New Experiment from Design Sample Execute Study, Collect “Sample Manifest” in Sample Manifest to validation of the Are the Assays handled in SAPIO? Is there any
Experiment workflow in in vivo Study or in vivo Experiment Name in Experiment, like title, Request from Sample to be analyzed to the Notify respective Study
Template in SAPIO Collection in ARIA Samples ARIA Sample Manager include in the Assay samples in the sample downstream workflow still in SAPIO?
SAPIO ARM in ARIA ARIA background, Manager Sample Assay Request Stakeholders
request list input
compounds, cell lines Quantification Assays
qPCR, Western Blot
MSD Genomics -
Vi-Cell Assay FACS Analysis
Flow Cytometry Illumina
NGS
CRISPR workflow
Design Sample
Collection and Record
Data in Studylog
Biomarker, targets Immunoblotting
MSD WES
Need a way to bring multiple
samples into system using a LC-MS
common ID Gyros
Barcode scanning
Cytokine Analysis
Collect Data files in common landing
-multiple samples for 1 project Nano string etc.
Send Samples to SARP, location DATA LAKE
-frost from freezer creates bad
scanning Bio Analysis Request
-tubes should have double ID to etc.
Ability to open a project page
help identify the sample. Other then
with drop down menu to
barcode Reported to DARE or
uploading files
-Location important to help identify
Easy to upload and store per IJC
the correct sample
assay type & instrument
You might also like
- Bug Hunter Methodology V4 (@jhaddix) : Finding SeedsDocument1 pageBug Hunter Methodology V4 (@jhaddix) : Finding SeedshsenNo ratings yet
- Parathyroid Glands: Serum PTH Levels Are Inappropriately Elevated For The LevelDocument4 pagesParathyroid Glands: Serum PTH Levels Are Inappropriately Elevated For The LevelNada MuchNo ratings yet
- 201 CraniosacralTherapyDocument65 pages201 CraniosacralTherapyUlisses Rodrigues Da Silva100% (1)
- Diphtheria Schematic Diagram (Pathophysiology0Document3 pagesDiphtheria Schematic Diagram (Pathophysiology0Kathlene Boleche100% (1)
- M-1 MCQDocument88 pagesM-1 MCQFrt TrfNo ratings yet
- PLSQL 2 3 PracticeDocument3 pagesPLSQL 2 3 PracticeSantiago Lara100% (1)
- Cervical Incompetence Case StudyDocument25 pagesCervical Incompetence Case StudyKavi rajputNo ratings yet
- Data Analysis in Next Generation SequencingDocument78 pagesData Analysis in Next Generation Sequencingparetini01No ratings yet
- PLSQL 2 3 Practice CompletaDocument3 pagesPLSQL 2 3 Practice CompletaEmanuel G.No ratings yet
- HESI Prep - Health AssessmentDocument248 pagesHESI Prep - Health Assessmentmeeeenon100% (4)
- 23 DC Acenocoumarol A Review PDFDocument6 pages23 DC Acenocoumarol A Review PDFnaga chaitanyaNo ratings yet
- System Design BlueprintDocument1 pageSystem Design BlueprintSathiamoorthy DuraisamyNo ratings yet
- Pattern Recognition Chip With 1024 Neurons in Parallel: Data SheetDocument2 pagesPattern Recognition Chip With 1024 Neurons in Parallel: Data Sheetteguh_setionoNo ratings yet
- Comparative Study of 80286,80386,80486, Pentium ProcessorsDocument4 pagesComparative Study of 80286,80386,80486, Pentium Processorsr_als100% (1)
- 2 Compiler - SlideDocument19 pages2 Compiler - Slidegilbertelena7898No ratings yet
- Aws Backup (Per 15 Minute) : Reverse FeedDocument1 pageAws Backup (Per 15 Minute) : Reverse FeedsiyakinNo ratings yet
- Csharp-Roadmap DrawioDocument1 pageCsharp-Roadmap Drawiobabaoarba0000No ratings yet
- Schweitzer Engineering Laboratories (SEL-735) Data Logging - MODBUS Registers MapDocument3 pagesSchweitzer Engineering Laboratories (SEL-735) Data Logging - MODBUS Registers MapAbzal KinayatovNo ratings yet
- RNA-seq Bioinformatics: Format and QC of Short ReadsDocument37 pagesRNA-seq Bioinformatics: Format and QC of Short ReadsManovriti ThakurNo ratings yet
- Block Diagram of Wyner-Ziv CODEC Implementation: TH Mt. ThreshDocument1 pageBlock Diagram of Wyner-Ziv CODEC Implementation: TH Mt. Threshapi-20005629No ratings yet
- All Tables DetailsDocument18 pagesAll Tables DetailsByron TejadaNo ratings yet
- ARM Cortex-A72 Block DiagramDocument1 pageARM Cortex-A72 Block Diagram文帅宋No ratings yet
- L22 Rout NupDocument6 pagesL22 Rout Nupsatyvan2003No ratings yet
- RFID at Metro GroupDocument26 pagesRFID at Metro Groupalbert_2mb100% (1)
- Book, Borrowers, Staff: Input Data Generate QR Code Generate Crystal ReportDocument2 pagesBook, Borrowers, Staff: Input Data Generate QR Code Generate Crystal ReportPaul GervacioNo ratings yet
- ARM Cortex-A12 Block DiagramDocument1 pageARM Cortex-A12 Block Diagram文帅宋No ratings yet
- 7 Text MiningDocument20 pages7 Text MiningmaskurNo ratings yet
- Bioinformatics Class1Document23 pagesBioinformatics Class1Satish SahuNo ratings yet
- Ifm Catalogue Control Systems GB 09Document212 pagesIfm Catalogue Control Systems GB 09ifm electronic100% (2)
- On Convolutional Precoding in PAC Codes: Mohammad Rowshan, Student Member, IEEE and Emanuele Viterbo, Fellow, IEEEDocument5 pagesOn Convolutional Precoding in PAC Codes: Mohammad Rowshan, Student Member, IEEE and Emanuele Viterbo, Fellow, IEEETudor MicuNo ratings yet
- FullertonCF Poster1 2Document1 pageFullertonCF Poster1 2mangkotiNo ratings yet
- LS/DS 3408: Quick Start GuideDocument2 pagesLS/DS 3408: Quick Start GuideDejanysNo ratings yet
- Mineral Identification in Seconds !!!Document2 pagesMineral Identification in Seconds !!!HAMCHI MohammedNo ratings yet
- ABI Prism 7000 Sequence Detection System User Guide - 2Document74 pagesABI Prism 7000 Sequence Detection System User Guide - 2Aqwin PolosoroNo ratings yet
- Inverse Folding Based Pre-Training For The Reliable Identification of Intrinsic Transcription Terminators - PLOS Computational BiologyDocument1 pageInverse Folding Based Pre-Training For The Reliable Identification of Intrinsic Transcription Terminators - PLOS Computational BiologyIndependz1No ratings yet
- Graph QLDocument1 pageGraph QLChukwuzube SamNo ratings yet
- Genomic DNA Libraries For Shotgun Sequencing ProjectsDocument40 pagesGenomic DNA Libraries For Shotgun Sequencing ProjectsGovind Kumar RaiNo ratings yet
- Introduction To LISP and VXLAN BRKRST-3045Document71 pagesIntroduction To LISP and VXLAN BRKRST-3045Harpreet Singh BatraNo ratings yet
- ARM Cortex-A57 Block DiagramDocument1 pageARM Cortex-A57 Block Diagram文帅宋No ratings yet
- Understanding OSPF Operation: Implement OSPF in The Service Provider NetworkDocument25 pagesUnderstanding OSPF Operation: Implement OSPF in The Service Provider NetworkAris HartonoNo ratings yet
- Acu-Expert TD EngDocument2 pagesAcu-Expert TD EngRadu BogdanNo ratings yet
- Building A Multi-Lingual OCR EngineDocument21 pagesBuilding A Multi-Lingual OCR EnginerossloveladyNo ratings yet
- Building A Multi-Lingual OCR EngineDocument21 pagesBuilding A Multi-Lingual OCR EngineYasserAl-mansourNo ratings yet
- 2020 - Identification of Library Functions Statically Linked To Linux Malware Without SymbolsDocument10 pages2020 - Identification of Library Functions Statically Linked To Linux Malware Without Symbolsaulia rachmaNo ratings yet
- ARM Cortex-A7 Core Block DiagramDocument1 pageARM Cortex-A7 Core Block Diagram文帅宋No ratings yet
- Computer ScienceDocument1 pageComputer SciencejayNo ratings yet
- DNA Seq AnalysisDocument28 pagesDNA Seq AnalysisArun VigneshNo ratings yet
- Cryptography Cheat SheetDocument3 pagesCryptography Cheat Sheetlbb1987No ratings yet
- RT PCR Gene Expression Intro v8Document1 pageRT PCR Gene Expression Intro v8ChristineDanNo ratings yet
- Arbol de Clase de Smalltalk - Rojas MoisesDocument1 pageArbol de Clase de Smalltalk - Rojas MoisesMoises RojasNo ratings yet
- DF - Sanpedro 2022 11 05 - 23 16Document4 pagesDF - Sanpedro 2022 11 05 - 23 16FRANCIS CASTILLO SANABRIANo ratings yet
- DF - Sanpedro 2022 11 05 - 23 17Document4 pagesDF - Sanpedro 2022 11 05 - 23 17FRANCIS CASTILLO SANABRIANo ratings yet
- List of Entitles in OOARSDocument5 pagesList of Entitles in OOARSKrushnavadan AcharyaNo ratings yet
- Tracing IP Address of Unidentified Persons in A Meeting SessionDocument4 pagesTracing IP Address of Unidentified Persons in A Meeting SessionTechnical MehmiNo ratings yet
- Novo PDocument24 pagesNovo Pfilipe guaranyNo ratings yet
- RustDocument1 pageRustLuann RodrigoNo ratings yet
- Preliminary Exam: DR Samuel Palermo Younghoon Song: A. Serial PRBS Generation and Error DetectionDocument27 pagesPreliminary Exam: DR Samuel Palermo Younghoon Song: A. Serial PRBS Generation and Error DetectionhoonigaNo ratings yet
- KCL NGScourse Session3 HandoutDocument13 pagesKCL NGScourse Session3 HandoutespartawithespartilhoNo ratings yet
- 09 Chapter4Document22 pages09 Chapter4Sravya ReddyNo ratings yet
- Flow ChartDocument2 pagesFlow ChartAgnathavasiNo ratings yet
- Retsol D 3030Document1 pageRetsol D 3030Naeemuddin yadgiriNo ratings yet
- PDF DocumentDocument19 pagesPDF DocumentMohamed aboalyNo ratings yet
- Brain Products GMBH - Products & Applications - Analyzer 2 PDFDocument3 pagesBrain Products GMBH - Products & Applications - Analyzer 2 PDFarun rayakwarNo ratings yet
- OAC 105.4 Selected New Features: Draft V2Document23 pagesOAC 105.4 Selected New Features: Draft V2nandakarsanNo ratings yet
- Rnaseq Workshop SlidesDocument110 pagesRnaseq Workshop SlidesFABIO ESTEBAN HERRERA ROCHANo ratings yet
- Database Programming With PL/SQL 2-3: Practice Activities: Recognizing Data TypesDocument3 pagesDatabase Programming With PL/SQL 2-3: Practice Activities: Recognizing Data TypesJust DuránNo ratings yet
- Study Data CaptureDocument2 pagesStudy Data Capturescribdusername_mooNo ratings yet
- Study Information Flow Mapping (In Vivo)Document3 pagesStudy Information Flow Mapping (In Vivo)scribdusername_mooNo ratings yet
- Study Information Flow MappingDocument2 pagesStudy Information Flow Mappingscribdusername_mooNo ratings yet
- Study Workflow - in Vivo ExperimentsDocument1 pageStudy Workflow - in Vivo Experimentsscribdusername_mooNo ratings yet
- A Blood Test For Early Cancer Detection Sparks DebateDocument6 pagesA Blood Test For Early Cancer Detection Sparks DebateGema Del Campo MontoyaNo ratings yet
- High Risk Case - Docx 1Document48 pagesHigh Risk Case - Docx 1MUNA NNo ratings yet
- Assisting in Endotracheal SuctioningDocument38 pagesAssisting in Endotracheal SuctioningMargarita Limon BalunesNo ratings yet
- Ichthyosis Vulgaris Atopic Dermatitis Asthma and AllergiesDocument2 pagesIchthyosis Vulgaris Atopic Dermatitis Asthma and Allergiesabortusprovocatus100% (1)
- Cardio SNAP-BNP NT-proBNP IdexxDocument4 pagesCardio SNAP-BNP NT-proBNP IdexxXenia FernandezNo ratings yet
- Thymic CystDocument6 pagesThymic CystNarendra BhattaraiNo ratings yet
- Cala InjuriesDocument6 pagesCala InjuriesMunashe HogweNo ratings yet
- Case-Study VincentDocument16 pagesCase-Study VincentKyle Audrie ArcalasNo ratings yet
- State Medical Faculty of West Bengal: 14C, Beliaghata Main Road, Kolkata-700085 : 2372 0185/2372 7183Document4 pagesState Medical Faculty of West Bengal: 14C, Beliaghata Main Road, Kolkata-700085 : 2372 0185/2372 7183riya RoyNo ratings yet
- BHRC Rates 01.04Document87 pagesBHRC Rates 01.04nishant00032No ratings yet
- Electrolyte Replacement Guide - ICU CliniciansDocument8 pagesElectrolyte Replacement Guide - ICU Cliniciansnolan4266No ratings yet
- Bipolar Handout 2Document12 pagesBipolar Handout 2hugoi_7No ratings yet
- Internal Medicine OSCE MindmapsDocument21 pagesInternal Medicine OSCE MindmapsCheru TadesseNo ratings yet
- Ante Partum and Intra Partum Fetal MonitoringDocument63 pagesAnte Partum and Intra Partum Fetal MonitoringasdfNo ratings yet
- A Minimal Dose Approach To Resistance Training For The Older AdultDocument7 pagesA Minimal Dose Approach To Resistance Training For The Older AdultThalesNo ratings yet
- Joker LineDocument10 pagesJoker LineVivekanand ChandrashekarNo ratings yet
- Arizona Communicable Disease FlipchartDocument98 pagesArizona Communicable Disease Flipchartapi-510866696No ratings yet
- Burns Summary of EvidenceDocument23 pagesBurns Summary of EvidenceYudhaNo ratings yet
- Small For Gestational Age: I. Definition/ DescriptionDocument11 pagesSmall For Gestational Age: I. Definition/ Descriptionfaye kimNo ratings yet
- Cholesterol Mortality Chart PDFDocument1 pageCholesterol Mortality Chart PDFTms Arn100% (1)
- VAD Patient Education ManualDocument12 pagesVAD Patient Education ManualIakovos DanielosNo ratings yet
- Immuno Hematology Blood TransfusionDocument1 pageImmuno Hematology Blood Transfusionudiptya_papai2007No ratings yet
- Adult Hip Pain (Ortho) WadaDocument25 pagesAdult Hip Pain (Ortho) WadaMeno AliNo ratings yet