Professional Documents
Culture Documents
Tutorial 6 Molecular Techniques
Uploaded by
Jason LeeOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Tutorial 6 Molecular Techniques
Uploaded by
Jason LeeCopyright:
Available Formats
MolBiol2C03: Genetics
Molecular techniques
Yusra Tariq, Jason Lee, Kriesha Eyer
Objectives:
Explain how could a fragment of DNA be amplified
Design PCR primers
Be able to differentiate between Sanger sequencing and Next Generation sequencing
Outline: In this week tutorial, you will discuss about PCR and design a set of primers for a given DNA
sequence.
Preparation
To prepare for the tutorial you should:
Read and bring to the tutorial this Guide
Read: 7.5 Methods of Molecular Genetic Analysis Make Use of DNA Replication Processes
(pg. 260 – 266) from “Genetics analysis - an integrated approach”
Polymerase Chain Reactions (PCR)
The polymerase chain reaction is a technique which amplifies a fragment of DNA a billion-fold in few
hours. The process consists of denaturing the double stranded DNA at 95 0C and replicating the DNA
using a DNA (Taq) polymerase enzyme.
The ingredients necessary for PCR are:
Template DNA – low amount of DNA from which we want to amplify a fragment of 100-5000 base
pairs (target sequence).
Primers – two small specific synthetic oligonucleotides, which exactly match two 15-30 unique base
pair region of the target sequence.
Taq polymerase – enzyme that catalyze the adding of new nucleotides to the 3’-OH of the primer.
Deoxynucleotide triphosphate (dNTPs) – equal mixture of dATP, dCTP, dGTP and dTTP. They are the
substrate for Taq polymerase.
Magnesium – required for Taq polymerase activity.
Buffer – to maintain the reaction’s pH.
Each PCR cycle has three steps1:
1. Denature. The PCR ingredients are heated at 95 0C for the double stranded DNA to denature into
single strands, by breaking the hydrogen bounds between the strands.
2. Annealing. The primers will bind (anneal) to the appropriate place on the template DNA that
contains the complementary sequence at around 550C. The forward primer will bind to the
bottom strand and the reverse primer will bind to the top strand.
3. Elongation. The Taq polymerase synthesis new DNA from 5’ to 3’, at 72 0C. DNA polymerase
adds dNTPs to the primers in the same direction as in cellular replication. At the end of the cycle
new two new DNA strands are produced.
The whole cycle is then repeated 25-40 times. With each cycle the amount of template DNA doubles,
which increases exponentially.
Designing primers1
Primers are crucial in the process of specific amplification of DNA. There are some rules you have to
follow when designing primers:
primers should be specific, each member of a primer pair anneals only to its target sequence in
the template DNA
primers should complement the template DNA;
primers should be 17-25 nucleotides in length;
the primers’ CG (G+C) content should be 40-60%;
primers should end (3') in a G or C, or CG or GC, this increases efficiency of annealing;
Melting temperatures (Tm) of hybrids between primers and their target sequence should be 55-
70oC are preferred;
3'-ends of primers should not be complementary (i.e. base pair), as otherwise primer dimers will
be synthesized preferentially to any other product;
primer self-complementarity (ability to form 2 o structures such as hairpins) should be avoided;
runs of four or more Cs or Gs at the 3'-ends of primers may promote mispriming at G or C-rich
sequences (because of stability of annealing), and should be avoided.
Melting temperature (Tm)1 is the temperature at which oligonucleotide primers dissociate from their
template. There are many formulas and computer programs to determine the theoretical melting
temperature. Tm is important for determining the annealing temperature (Ta), which is the optimal
temperature for the primers to anneal to target DNA.
In general Ta is 3-50C lower than the calculated Tm. If the annealing temperature is too high, the
primers anneal poorly to the template and the yield of amplified DNA is very low. If the annealing
temperature is too low, the primers will anneal nonspecific, resulting in the amplification of
unwanted segments of DNA.
1. Adapted from “Molecular Cloning: A Laboratory Manual”, by Joe Sambrook (2001).
MolBiol2C03: Genetics
Molecular techniques
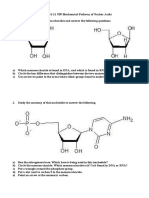
1. Which of these can DNA polymerase add on to?
A) 5’----------------------------------3'
3’----------------------------------5’
B) 5’-------------------3’
3’----------------------------------5’
C) 5’----------------------------------3’
3’-------------------5’
2. Given a sequence of double stranded DNA, name the primers, both forward and reverse that would
anneal to the sequence (only extend the primers for 6 bases).
5’-AAGTCGTAACC TTGCACGGTAGC-3’
3’-T TCAGCATTGGAACGTGCCATCG-5’
Forward Reverse
A) 5’-AACCTT-3’ 5’-AAGTCG-3’
B) 5’-TTCAGC-3’ 5’-CGGGGC-3’
C) 5’-AAGTCG-3’ 5’-GCTACC-3’
D) 5’-GGTAGC-3’ 5’-GCCCCG-3’
E) 5’-ATGCGT-3' 5’-TTGGAA-3'
3. You are working for 23andme and you have to determine your clients’ genotype for SNP rs2472297.
This SNP is a point mutation of CYP1A2 gene, involved in coffee metabolism. The T allele of rs2472297 is
consistently linked to a higher coffee intake than the ancestral C allele.
First you want to amplify a ~ 200bp fragment that includes the SNP, and for this you need to design
primers.
5’-TTGCACAAA TGTTTCCCAA GCCAATGAAT TATGAAATAA GCCTTGCAGG GGAATGTTTG AGCTACTGAA
GGGCATGTTC GGCATGATCA GAGGAACACT CTGTTTCCAA GTTAGTA ATG Y* CTCTTTACAT ATAAACACTG
ATGTTTGTTG ATAATGAACA TTCATGTCCT CAATCAAAGC AGAGCCTCTG GGGACACTTC TCTATCAGCC
AAGTGCTCCT AGAAGTGTTC-3’
*where Y represents the SNP and it could be C or T
a) Underline and write your primers in a 5’-3’ orientation.
Forward: 5’ – TTGCACAAATGTTTCCCAAG – 3’
Reverse: 5’ – GAACACTTCTAGGAGCACTT – 3'
b) What is their annealing temperature using the Wallace/Ikatura rule?
Tm forward =2 o (number of A's and T's) + 4 o (number of G's and C's)
= 2 o (12) + 4 o (8)
= 56 o C
Ta forward = Tm-5 o
= 56-5
= 51 o C
Tm reverse = 2 o (number of A's and T's) + 4 o (number of G's and C's)
= 2 o (11) + 4 o (9)
= 58 degrees
Ta reverse = Tm - 5 o
= 58-5
= 53 o C
You might also like
- DNA Methods in Food Safety: Molecular Typing of Foodborne and Waterborne Bacterial PathogensFrom EverandDNA Methods in Food Safety: Molecular Typing of Foodborne and Waterborne Bacterial PathogensNo ratings yet
- Principle of The PCR: This Technique Is Invented in 1984 by Kary Mulis. The Purpose of A PCRDocument11 pagesPrinciple of The PCR: This Technique Is Invented in 1984 by Kary Mulis. The Purpose of A PCRantesar reheemNo ratings yet
- Prerequisite of A PCR: Logarithmic and Controlled FashionDocument18 pagesPrerequisite of A PCR: Logarithmic and Controlled FashionlkokodkodNo ratings yet
- Next Generation Sequencing and Sequence Assembly: Methodologies and AlgorithmsFrom EverandNext Generation Sequencing and Sequence Assembly: Methodologies and AlgorithmsNo ratings yet
- Maria Afzal M.phill-03 Submitted To: Prof. Dr. Farkhanda JabeenDocument44 pagesMaria Afzal M.phill-03 Submitted To: Prof. Dr. Farkhanda JabeenMuhammad MushtaqNo ratings yet
- Lab Manual 3Document7 pagesLab Manual 3syazaismailNo ratings yet
- PCR and Agarose Gel ElectrophoresisDocument6 pagesPCR and Agarose Gel ElectrophoresisÍrisNo ratings yet
- Polymerase Chain Reaction (PCR)Document37 pagesPolymerase Chain Reaction (PCR)Haleema SultanNo ratings yet
- PCR An OverviewDocument41 pagesPCR An OverviewnassradabourNo ratings yet
- The Olymerase Hain Eaction: Zhang-Haifeng Department of BiochemistryDocument42 pagesThe Olymerase Hain Eaction: Zhang-Haifeng Department of Biochemistryapi-19916399No ratings yet
- The Polymerase Chain Reaction and Sequence-Tagged SitesDocument7 pagesThe Polymerase Chain Reaction and Sequence-Tagged SitesPisi MicaNo ratings yet
- General PCR Guidelines: PrimersDocument13 pagesGeneral PCR Guidelines: PrimersAdi NugrahaNo ratings yet
- 2019-20 Bioinformatics CDocument28 pages2019-20 Bioinformatics CbeaNo ratings yet
- The Polymerase Chain Reaction (PCR)Document50 pagesThe Polymerase Chain Reaction (PCR)Deana NamirembeNo ratings yet
- 4.PCR Primer Designing (Deepa Chouhan) - 1Document14 pages4.PCR Primer Designing (Deepa Chouhan) - 1VK 360 CREATIONSNo ratings yet
- Polymerase Chain Reaction (PCR)Document3 pagesPolymerase Chain Reaction (PCR)biotech_vidhyaNo ratings yet
- Full-Length cDNA Cloning and Determination of mRNA 5 and 3 Ends by Amplification of Adaptor-Ligated cDNADocument9 pagesFull-Length cDNA Cloning and Determination of mRNA 5 and 3 Ends by Amplification of Adaptor-Ligated cDNADiksha GahlotNo ratings yet
- PCR AmplificationDocument5 pagesPCR AmplificationNabiha KsNo ratings yet
- Polymerase Chain ReactionDocument34 pagesPolymerase Chain Reactionmokshgoyal2597100% (3)
- Polymerase Chain Reaction 1Document8 pagesPolymerase Chain Reaction 1Ye Htut AungNo ratings yet
- MKBS221. PRAC 5 REPORT Last OneDocument11 pagesMKBS221. PRAC 5 REPORT Last OneNOMKHULEKO ALICENo ratings yet
- DNA/RNA Pathway AnalysisDocument6 pagesDNA/RNA Pathway AnalysisBrian Chi Yan Cheng 鄭智仁No ratings yet
- PCRDocument42 pagesPCRNopiyana PujiastutiNo ratings yet
- Quiz #3: Biochemical Engineering Fall 2003Document5 pagesQuiz #3: Biochemical Engineering Fall 2003Princess Janine CatralNo ratings yet
- Primer Design ExerciseDocument34 pagesPrimer Design ExerciseRita karen Pacheco CabañasNo ratings yet
- PCR ManualDocument6 pagesPCR ManualjiaNo ratings yet
- PCR Primer DesignDocument13 pagesPCR Primer DesignAiman ArshadNo ratings yet
- PCR Overview GoldBioDocument1 pagePCR Overview GoldBioKIARAH EUNIZE GICANo ratings yet
- Chapter 5 - PrimerDesign and Restriction Digestion Analysis PRINTDocument112 pagesChapter 5 - PrimerDesign and Restriction Digestion Analysis PRINTshuma chemedaNo ratings yet
- Primer Designing For PCRDocument13 pagesPrimer Designing For PCRiftikharsubha5No ratings yet
- Adv 4.Document18 pagesAdv 4.harseen rahimNo ratings yet
- Polymerase Chain Reaction (PCR)Document17 pagesPolymerase Chain Reaction (PCR)pushpavalliNo ratings yet
- Primer Design 2013 PDFDocument58 pagesPrimer Design 2013 PDFMaila EscuderoNo ratings yet
- Primer Designing For PCR: ColloquiumDocument3 pagesPrimer Designing For PCR: ColloquiumSakshi SangleNo ratings yet
- BENGAL SCHOOL OF TECHNOLOGY STUDENT ANKUSH CHAKRABORTY'S ADVANCED PHARMACEUTICAL BIOTECHNOLOGY CLASS PROJECT ON POLYMERASE CHAIN REACTIONDocument13 pagesBENGAL SCHOOL OF TECHNOLOGY STUDENT ANKUSH CHAKRABORTY'S ADVANCED PHARMACEUTICAL BIOTECHNOLOGY CLASS PROJECT ON POLYMERASE CHAIN REACTIONAnkush ChakrabortyNo ratings yet
- PCR Amplification Lab ReportDocument5 pagesPCR Amplification Lab ReportWilson Chan100% (7)
- DNA amplification 2020Document5 pagesDNA amplification 2020Blameless ArikoNo ratings yet
- How To Design PrimersDocument14 pagesHow To Design PrimersDeepak RawoolNo ratings yet
- TaKaRa Successful PCR Guide 3rd EdDocument60 pagesTaKaRa Successful PCR Guide 3rd EdSali GiftNo ratings yet
- PCR Amplification of DNA SequencesDocument56 pagesPCR Amplification of DNA SequencesSamiksha SharmaNo ratings yet
- Polymerase Chain ReactionDocument27 pagesPolymerase Chain ReactionundalliNo ratings yet
- Molecular Diagnostics Techniques Polymerase Chain ReactionDocument7 pagesMolecular Diagnostics Techniques Polymerase Chain ReactionJamesNo ratings yet
- Igem Training 1Document45 pagesIgem Training 1api-343562611No ratings yet
- DNA Techniques (PCR and Sequencing)Document4 pagesDNA Techniques (PCR and Sequencing)ffarrugia12No ratings yet
- Chapter8 PrimerdesigningDocument8 pagesChapter8 PrimerdesigningSakshi SangleNo ratings yet
- Primer Design: (In This Case The Word Primer Means An Oligonucleotide)Document19 pagesPrimer Design: (In This Case The Word Primer Means An Oligonucleotide)kundansagarNo ratings yet
- PCR Fundamentals: Understanding the Technique and Optimizing Reaction ConditionsDocument5 pagesPCR Fundamentals: Understanding the Technique and Optimizing Reaction ConditionsOh RoopzNo ratings yet
- PCRDocument34 pagesPCRSumitNo ratings yet
- PCR - Polymerase Chain ReactionDocument37 pagesPCR - Polymerase Chain ReactionNandana Nayana Kumara100% (4)
- Polymerase Chain Reaction (PCR)Document37 pagesPolymerase Chain Reaction (PCR)PRUDHVIAZADNo ratings yet
- Morgan 1922: 2000 Genes in 4 Chromosome of D.: MelanogasterDocument44 pagesMorgan 1922: 2000 Genes in 4 Chromosome of D.: MelanogasterlordniklausNo ratings yet
- Lorenz2012 PDFDocument15 pagesLorenz2012 PDFCatherine Cinthya RIMAC PANEZNo ratings yet
- Successful PCR Guide: 3rd EditionDocument60 pagesSuccessful PCR Guide: 3rd Editionlpr121618100% (2)
- Understanding PCR: A Guide to Polymerase Chain ReactionDocument23 pagesUnderstanding PCR: A Guide to Polymerase Chain ReactionIkrimah FithriyandiniNo ratings yet
- Race PCR PDFDocument6 pagesRace PCR PDFmanuel1788No ratings yet
- Tips For Molecular TechniquesDocument14 pagesTips For Molecular TechniquesSara BibiNo ratings yet
- PCR-RFLP Untuk Deteksi Polimorfisme DNA 2020Document23 pagesPCR-RFLP Untuk Deteksi Polimorfisme DNA 2020Raymond Yehezkiel TambunNo ratings yet
- Polymerase Chain ReactionDocument14 pagesPolymerase Chain ReactionwiraNo ratings yet
- Polyme Rase Chain Reactio N: Course Code: 501Document23 pagesPolyme Rase Chain Reactio N: Course Code: 501Humayun ArshadNo ratings yet
- Chapter 5 Lesson 1 Genetically Modified OrganismsDocument7 pagesChapter 5 Lesson 1 Genetically Modified Organismsdel143masNo ratings yet
- Pgapz Manual InvitrogenDocument44 pagesPgapz Manual InvitrogenHeny RHNo ratings yet
- Bio SensorsDocument10 pagesBio SensorsDipti GNo ratings yet
- Bacteria vs Virus: Key Differences and How Pathogenic Bacteria Harm the Human BodyDocument11 pagesBacteria vs Virus: Key Differences and How Pathogenic Bacteria Harm the Human BodySakib Rahman DiptoNo ratings yet
- Introduction Micro SallyDocument18 pagesIntroduction Micro Sallygosallyfirst9771No ratings yet
- Megakaryopoiesis and Thrombopoiesis GuideDocument2 pagesMegakaryopoiesis and Thrombopoiesis GuideIberisNo ratings yet
- Introduction To Differential Gene Expression Analysis Using RNA-seqDocument97 pagesIntroduction To Differential Gene Expression Analysis Using RNA-seqJimmy DazaNo ratings yet
- Allelic diversity for salt stress genes in Indian riceDocument9 pagesAllelic diversity for salt stress genes in Indian ricesalma sabilaNo ratings yet
- RocheDocument0 pagesRocheIrma KurniawatiNo ratings yet
- Nucleic Acids WorksheetDocument4 pagesNucleic Acids WorksheetNatalie PembertonNo ratings yet
- Origins of Life LabDocument4 pagesOrigins of Life LabJoey Ma50% (2)
- Fda 21 CFR 820 PDFDocument2 pagesFda 21 CFR 820 PDFAmber0% (2)
- Sel HewanDocument489 pagesSel HewanRahmat Reza100% (1)
- People V UmanitoDocument6 pagesPeople V UmanitoLilu BalgosNo ratings yet
- Flashcards MicrobiologyDocument26 pagesFlashcards MicrobiologyabhishekNo ratings yet
- Southwestern BlottingDocument3 pagesSouthwestern Blottingheran_1006No ratings yet
- Test Bank For Principles of Biochemistry 4th Edition HortonDocument12 pagesTest Bank For Principles of Biochemistry 4th Edition HortonSandra Hornyak100% (8)
- CV - Munvar M ShaikDocument5 pagesCV - Munvar M ShaikMohammad GouseNo ratings yet
- Ch. 23 Lecture Outline PDFDocument15 pagesCh. 23 Lecture Outline PDFCSERM UNASNo ratings yet
- 5 Factors Affecting Morphogenesis in VitroDocument20 pages5 Factors Affecting Morphogenesis in VitroAlec Liu0% (3)
- Biomolecules Recap Answer Key by The Amoeba Sisters AnswerkeyDocument3 pagesBiomolecules Recap Answer Key by The Amoeba Sisters AnswerkeySandra Lopez50% (2)
- General Biology 1 Long TestDocument3 pagesGeneral Biology 1 Long TestQueng Eledia100% (1)
- B. Sc. (Hons) Botany 4th Semester-2022-MergedDocument44 pagesB. Sc. (Hons) Botany 4th Semester-2022-MergedAnsh KumarNo ratings yet
- 120-202 Fall 21 Lab ManualDocument104 pages120-202 Fall 21 Lab ManualLeslie NunezNo ratings yet
- 2021-22 6th SEM PYQ - MergedDocument7 pages2021-22 6th SEM PYQ - MergedPriyanshu UpadhyayNo ratings yet
- B InggrisDocument8 pagesB InggrisTrisna EniNo ratings yet
- IMNCIDocument13 pagesIMNCIJayalakshmiullasNo ratings yet
- Hybrid Compounds As Direct Multitarget Ligands: A ReviewDocument36 pagesHybrid Compounds As Direct Multitarget Ligands: A ReviewJames TerryNo ratings yet
- Virus BioinformaticsDocument332 pagesVirus BioinformaticsVictor Delgado PNo ratings yet
- 9th Class Biology Full Book TestDocument2 pages9th Class Biology Full Book TestSaqib Ali Bhutta80% (5)