Professional Documents
Culture Documents
Cmci Poster Final
Cmci Poster Final
Uploaded by
api-559032517Original Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Cmci Poster Final
Cmci Poster Final
Uploaded by
api-559032517Copyright:
Available Formats
Keeping It All Together: 2 Projects Testing Stability Predictions in a Virus
Adrienne E. Fairbanks,1 Natalya Usachenko,2
LuAnn Scott, Emma R. Altman, James T. Van Leuven,
Jagdish Patel, Craig R. Miller and Holly A. Wichman
Department of Biological Sciences & the Center for Modeling Complex Interactions, University of Idaho, Moscow Idaho
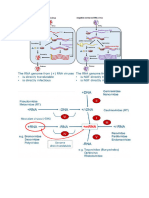
2Testing the Capacity to Over-Stabilize 1Testing Metrics of Transcript Stability
the Viral Capsid .
What is the relationship between synonymous
Can a virus be over-stabilized through the alteration of mutations engineered to increase or decrease viral
its minor capsid protein? transcript stability and fitness?
Hypothesis Metrics
CAI Codon Adaptation Index
Combination of two or more stabilizing mutations in a viral capsid protein will have
CPB Codon Pair Bias
an additive effect on the overall stability of the protein, potentially reducing viral
sCAI Starvation CAI
fitness in two ways:
tAI tRNA Adaptation Index
1. Making capsid assembly more difficult and thereby reducing the number of
ITE Index of Translation Elongation
viable offspring.
SD Shine-Delgarno
2. Increasing the overall stability of the capsid itself, reducing its ability to infect
mFE 5’ mRNA Folding Energy
a host cell.
Bacteriophage φX174 TPI tRNA Pairing Index
Experimental Design An efficient virus model
• Combine stabilizing mutations in protein G of φX174, the minor capsid/major Proposed Experiment
spike protein (Fig.1). • Non-pathogenic
• Choose 5 phage each with a single amino acid mutation predicted to stabilize
• Design 4 high & 4 low stability mutants for genes F, G, & H, major
• Small genome size
binding of the protein (Fig. 2). capsid, major spike, and pilot proteins, respectively (Fig. 4)
• Large populations
• Add each amino acid mutation to each of the other four phage by site directed • Have gene fragments synthesized & cloned into plasmids
• Ease of manipulation of genome sequence
mutagenesis (Fig. 3). • Use assembly platform to create mutant phage genomes (Fig. 5)
o Site directed mutagenesis
• Assess the change in fitness – measured by growth rate or plaque size. • Transform into host cells, pick plaques and sequence
o Engineered fragments inserted using assembly platform
• Perform fitness assays
A B C
• Assays available for fitness, attachment, thermal stability, burst
Figure 2. FoldX time, burst size, length of eclipse, length of assembly, rate of
predictions for changes
in stability for binding (x assembly and mortality rate
axis) and folding (y
axis) when replacing
the wild type amino acid
Overarching Context
at A) G11, B) G4, C)
G45, D) G128, and E)
Understanding basic mechanisms of viral adaptation
G117. The predicted
D E
∆∆G (change from wt Project 1
change in free energy) Figure 4. The φX174 genome is a single, circular chromosome of 5386 bp and contains 11
for the 19 other amino • This research is part of an effort to develop a genes, some overlapping. Promoters and terminators for the transcripts are below and the cloned
acids is represented by “recipe” for designing viral vaccines. assembly fragments that will be modified in these studies are above.
dots. Negative values • Synonymous mutations could be one
are stabilizing.
ingredient
• But…synonymous mutations affect more than
Figure 3. Location of
just codons
stability mutations to be A B
combined. A) A monomer of
G (light blue) and a Project 2
monomer of F (purple)
shown in relationship to the • Protein stability – important determining factor in
pentamers of G/F (grey). B) protein evolution. Figure 5. Assembly platform for φX174. A) 14 segments of phage genome (black), flanked by
Ribbon structure of protein
G with the locations of the • Used in predictive modeling. (Doore et al.). BsmB1 restriction sites, cloned into a pCRII-TOPO plasmid (grey). One or more fragments is
mutated (gold). B) Phage inserts are excised with BsmB1, which cuts in the phage sequence.
five stability mutants shown • Increased capsid stability previously observed in C) Inserts are ligated together using the unique 4 bp sticky ends created by BsmB1 to
in dark blue. high temperature phage mutants with higher fitness reconstruct the genome with the incorporation of the mutant fragment(s).
than wild type (Lee et al.).
Current Results • Correlations exist between viral evolution and Expected Results
climate change; this study will continue to explore
• Viable double mutants in two phage so far: G4_M/117_M, • Reduced fitness in all 24 phages caused by reduced numbers of
this phenomenon.
G128_V/117_M. protein
• Sequencing and fitness assessment ongoing. o Stabilizing mutations slow down translation
• Plan to add a third stabilizing mutation to all viable doubles. Acknowledgements o Destabilizing mutations cause degradation of mRNA faster than
References Phage image by Ben Darby usual
Doore, Sarah M., Fane, Bentley A. (2016) “The microviridae: Diversity, assembly, and experimental evolution”. Virology vol 491, p. 45-55
Lee KH, Miller CR, Nagel AC, Wichman HA, Joyce P, et al. (2011) “First-Step Mutations for Adaptation at Elevated Temperature Increase Capsid
Stability in a Virus”. PLoS ONE 6(9): e25640
Funding provided by NSF EPSCoR grant OIA-1736253, NIH COBRE grant • Modelers will evaluate results to better understand the relationship
McKenna, R., L. L. Ilag, and M. G. Rossmann. 1994. Analysis of the single-stranded DNA bacteriophage phi X174, refined at a resolution of 3.0 A. J P20GM104420 and NIH R01 grant GM076040. between the metrics and synonymous mutations
Mol Biol 237:517-543.
You might also like
- Food and NutritionDocument36 pagesFood and NutritionM.g. Alomia67% (3)
- Evidence-Informed Primary Care Management of Low Back Pain - Clinical Practice Guideline - CanadaDocument49 pagesEvidence-Informed Primary Care Management of Low Back Pain - Clinical Practice Guideline - CanadaCambriaChicoNo ratings yet
- 12 Weeks To Your Hottest Body EverDocument49 pages12 Weeks To Your Hottest Body EverSixp8ck100% (1)
- Introduction To Gerontology and Theories of AgingDocument106 pagesIntroduction To Gerontology and Theories of AgingCyden Shame delos Santos100% (1)
- Cultivation of White Button MushroomDocument55 pagesCultivation of White Button MushroomKaran R100% (6)
- Professor Chamberlains 10 Rules of Normal ECGDocument12 pagesProfessor Chamberlains 10 Rules of Normal ECGAnusha Verghese83% (6)
- Relaksasi Genggam JariDocument13 pagesRelaksasi Genggam JariIan ClaxNo ratings yet
- Science 2019 366 6469 TwisDocument5 pagesScience 2019 366 6469 TwisAnderson Widmer Morales VillarrealNo ratings yet
- Duplo Mutante Do Inibidor de Quimotripsina 2 Estabilizado Através Do Aumento Da Entropia ConformacionalDocument11 pagesDuplo Mutante Do Inibidor de Quimotripsina 2 Estabilizado Através Do Aumento Da Entropia ConformacionalRenan Guilherme de Oliveira GuihNo ratings yet
- siRNA - ApplicationsDocument7 pagessiRNA - ApplicationsMahmood-S ChoudheryNo ratings yet
- Cloning A Vaccinia Virus Host Range Determinant, C7L, Into A Bacterial Expression Vector For Biophysical Analysis of The Purified ProteinDocument1 pageCloning A Vaccinia Virus Host Range Determinant, C7L, Into A Bacterial Expression Vector For Biophysical Analysis of The Purified ProteinJennifer ChenNo ratings yet
- Caracterização Do Complexo Fuzzy de Alta Afinidade Entre o Domínio Desordenado Da Oncoproteína E7 Do HPV de Alto Risco e o Domínio TAZ2 Do CBPDocument12 pagesCaracterização Do Complexo Fuzzy de Alta Afinidade Entre o Domínio Desordenado Da Oncoproteína E7 Do HPV de Alto Risco e o Domínio TAZ2 Do CBPRenan Guilherme de Oliveira GuihNo ratings yet
- AAV Vector Development, Production and Application IIDocument1 pageAAV Vector Development, Production and Application IIrohitgazmer56No ratings yet
- Aptamer-Based Electrochemical Biosensor For Interferon Gamma DetectionDocument6 pagesAptamer-Based Electrochemical Biosensor For Interferon Gamma DetectionRenan Guilherme de Oliveira GuihNo ratings yet
- The Hepatitis B Virus Core Protein Intradimer Interface Modulates Capsid Assembly and StabilityDocument9 pagesThe Hepatitis B Virus Core Protein Intradimer Interface Modulates Capsid Assembly and StabilityMarius StancuNo ratings yet
- Novagen Competent Cells BrochureDocument12 pagesNovagen Competent Cells BrochureMariaNo ratings yet
- Seminar - SAMADocument30 pagesSeminar - SAMAsamaNo ratings yet
- Crystal Structure of An in Vitro Affinity-And Specificity-Matured Anti-Testosterone Fab in Complex With TestosteroneDocument7 pagesCrystal Structure of An in Vitro Affinity-And Specificity-Matured Anti-Testosterone Fab in Complex With Testosterone彭凯臣No ratings yet
- Summary ShortDocument10 pagesSummary ShortDelon van den AkkerNo ratings yet
- Notes For VDJ ArrangementDocument4 pagesNotes For VDJ ArrangementShane FernandezNo ratings yet
- Lodish CloningDocument38 pagesLodish CloninggustavoNo ratings yet
- Deepmind's Ai Predicts Structures For A Vast Trove of ProteinsDocument1 pageDeepmind's Ai Predicts Structures For A Vast Trove of ProteinsLuis RomeroNo ratings yet
- Artikel EBPDocument2 pagesArtikel EBPWahyu NugraheniNo ratings yet
- 3 Lec Secretory Folding HODocument11 pages3 Lec Secretory Folding HOHamud ShmalyNo ratings yet
- Antibody LandscapeDocument10 pagesAntibody LandscapeRajkumar PaulNo ratings yet
- Targeted Protein Degradation - Expanding The ToolboxDocument15 pagesTargeted Protein Degradation - Expanding The ToolboxAndonis AngelovNo ratings yet
- Pre-mRNA Splicing Life at The Centre of The Central DogmaDocument3 pagesPre-mRNA Splicing Life at The Centre of The Central DogmaQwyn Kym De GuzmanNo ratings yet
- The Molecular Basis of GPCR Activation - Weis and Kobilka, 2018Document23 pagesThe Molecular Basis of GPCR Activation - Weis and Kobilka, 2018Cynthia RamírezNo ratings yet
- CHO HCP Poster Presented at Bangalore India Bio2011Document1 pageCHO HCP Poster Presented at Bangalore India Bio2011mercerexpNo ratings yet
- HindiDocument5 pagesHindiharshita nairNo ratings yet
- Supplementary Material: 3. Virtual Laboratories: Access InformationDocument11 pagesSupplementary Material: 3. Virtual Laboratories: Access InformationRocio MontanoNo ratings yet
- Light-Inducible Recombinases For Bacterial Optogenetics: AccessDocument9 pagesLight-Inducible Recombinases For Bacterial Optogenetics: AccessDuxan Arancibia RadichNo ratings yet
- Blank CU, Haanen JB, Ribas A, Et Al. The Cancer Immunogram. ScienceDocument4 pagesBlank CU, Haanen JB, Ribas A, Et Al. The Cancer Immunogram. Sciencedaniela francoNo ratings yet
- Lecture 4 - Structure of VirusesDocument57 pagesLecture 4 - Structure of VirusesERNEST GABRIEL ADVINCULANo ratings yet
- Tegm - FinalDocument33 pagesTegm - FinalDani GutiérrezNo ratings yet
- Unraveling The Hook EffectDocument9 pagesUnraveling The Hook EffectJoana BarbosaNo ratings yet
- Labster Reviewer FinalsDocument6 pagesLabster Reviewer Finalscharles babasaNo ratings yet
- The Bacterial RecA Protein and Recombinationlal Dna Repair of Stalled Replication ForksDocument32 pagesThe Bacterial RecA Protein and Recombinationlal Dna Repair of Stalled Replication ForksHaru SahaNo ratings yet
- Khalif Eh Zadeh 2020Document13 pagesKhalif Eh Zadeh 2020Caroldemort :3No ratings yet
- Innate Lymphoid Cells To The Rescue: One Gene For Three Calcium CurrentsDocument3 pagesInnate Lymphoid Cells To The Rescue: One Gene For Three Calcium CurrentsElizabeth ParsonsNo ratings yet
- 1 s2.0 S1050464821001996 MainDocument13 pages1 s2.0 S1050464821001996 MainKevin MaiseyNo ratings yet
- 116 4thjuly2012 IsThereHiggsParticleDocument1 page116 4thjuly2012 IsThereHiggsParticleSubramanian AnanthanarayananNo ratings yet
- Dan R T CRISPE-2Document33 pagesDan R T CRISPE-2mouid0.2003No ratings yet
- Retroviral Mediated Gene Transfer: RetrovirusDocument6 pagesRetroviral Mediated Gene Transfer: RetrovirusDhaval ShahNo ratings yet
- Grupo 2 Miercoles 2022Document3 pagesGrupo 2 Miercoles 2022Pamela Francisca Mamani ViamonteNo ratings yet
- 유전체학Document19 pages유전체학wiwaxia1911No ratings yet
- NRDD The Landscape For Lipid Nanoparticle Based Genomic Medicines PDFDocument4 pagesNRDD The Landscape For Lipid Nanoparticle Based Genomic Medicines PDFru ruNo ratings yet
- zhang et al 2022 protac degrader of estrogen receptor α targeting dna binding domain in breast cancerDocument10 pageszhang et al 2022 protac degrader of estrogen receptor α targeting dna binding domain in breast cancernokikufubun72No ratings yet
- B InggrisDocument9 pagesB InggrisRizma DwiNo ratings yet
- Gene MutationsDocument7 pagesGene MutationsManan GhoshNo ratings yet
- DNA Clonning: Done By: Enid Hii Lin Wei & Chong Hui JunDocument16 pagesDNA Clonning: Done By: Enid Hii Lin Wei & Chong Hui JunHui JunNo ratings yet
- Mini Review 2Document10 pagesMini Review 2Aleena KhanNo ratings yet
- Crescimento Dependente de Peptídeo em Levedura Através Da Expressão Genética Essencial Ativada Por Peptídeo FinoGPCRDocument10 pagesCrescimento Dependente de Peptídeo em Levedura Através Da Expressão Genética Essencial Ativada Por Peptídeo FinoGPCRRenan Guilherme de Oliveira GuihNo ratings yet
- Science 8 Vertical AlignmentDocument4 pagesScience 8 Vertical AlignmentHenno Nickole Vince A. BugtongNo ratings yet
- Anti-Interferon Beta: Certificate of AnalysisDocument2 pagesAnti-Interferon Beta: Certificate of AnalysisyolsuzzNo ratings yet
- Biotech GenDocument33 pagesBiotech GenSashaNo ratings yet
- Breakthrough To Genome Editing: EditorialDocument1 pageBreakthrough To Genome Editing: EditorialMauricio RíosNo ratings yet
- Development of Transgene Construct Using Coat Protein Gene of Papaya Ringspot Virus and Its ValidationDocument6 pagesDevelopment of Transgene Construct Using Coat Protein Gene of Papaya Ringspot Virus and Its ValidationMathew UsfNo ratings yet
- Ivermectin Promotes Peripheral Nerve RegenerationDocument11 pagesIvermectin Promotes Peripheral Nerve RegenerationbiaravankNo ratings yet
- Mehmet Emin Uslu: New Biotechnology Volume 29S September 2012Document1 pageMehmet Emin Uslu: New Biotechnology Volume 29S September 2012waleedNo ratings yet
- Characterization of Expressed Genes in Kidney Cells of Japanese Flounder Paralichthys Olivaceus Following Treatment With Cona/Pma and LpsDocument8 pagesCharacterization of Expressed Genes in Kidney Cells of Japanese Flounder Paralichthys Olivaceus Following Treatment With Cona/Pma and LpsWilliam RamirezNo ratings yet
- Guthertz Et Al 2022 The Effect of Mutation On An Aggregation Prone Protein An in Vivo in Vitro and in Silico AnalysisDocument11 pagesGuthertz Et Al 2022 The Effect of Mutation On An Aggregation Prone Protein An in Vivo in Vitro and in Silico AnalysisrinjaniNo ratings yet
- PosterDocument1 pagePosterapi-509694663No ratings yet
- Cell Junction by T. DevasenaDocument19 pagesCell Junction by T. DevasenaLeslie RovoNo ratings yet
- Cur Map Grade 10 q3 2Document3 pagesCur Map Grade 10 q3 2Malicah MamaNo ratings yet
- Milinski y Rockenbach - Spying On Others EvolvesDocument3 pagesMilinski y Rockenbach - Spying On Others EvolvesMateo MontoyaNo ratings yet
- Usachenko CV EportfolioDocument4 pagesUsachenko CV Eportfolioapi-559032517No ratings yet
- Eng317project1 MemoreportDocument2 pagesEng317project1 Memoreportapi-559032517No ratings yet
- Usage of MicropipetteDocument1 pageUsage of Micropipetteapi-559032517No ratings yet
- Google Sheets Vs Microsoft Excel: Introduction ... P1Document11 pagesGoogle Sheets Vs Microsoft Excel: Introduction ... P1api-559032517No ratings yet
- Carbonateprojectresearchposter UpdatedDocument1 pageCarbonateprojectresearchposter Updatedapi-559032517No ratings yet
- Eng317infographic PDFDocument1 pageEng317infographic PDFapi-559032517No ratings yet
- Group 6 Chapter 1 3 1 AqumarineDocument15 pagesGroup 6 Chapter 1 3 1 AqumarineMark D-weh ReloxNo ratings yet
- HPV Testing in The Follow-Up of Women Post Colposcopy Treatment - Final VersionDocument16 pagesHPV Testing in The Follow-Up of Women Post Colposcopy Treatment - Final VersionPaulo César López BarrientosNo ratings yet
- Caregiver Benefits BrochureDocument8 pagesCaregiver Benefits BrochureNiña MoradaNo ratings yet
- The Pathogenesis and Prevention of Corneal Graft Melting After KeratoplastyDocument9 pagesThe Pathogenesis and Prevention of Corneal Graft Melting After KeratoplastywahyuNo ratings yet
- Tamoxifen CitrateDocument3 pagesTamoxifen Citrateapi-3797941No ratings yet
- ACLS Tachycardia Algorithm For Managing Stable TachycardiaDocument4 pagesACLS Tachycardia Algorithm For Managing Stable TachycardiaizkalotlNo ratings yet
- 8-17-20 Protocols On Voter Regs (Revised As of August 12, 2020)Document8 pages8-17-20 Protocols On Voter Regs (Revised As of August 12, 2020)DaveKarlRamada-MaraonNo ratings yet
- Cordyceps 16mayvjDocument2 pagesCordyceps 16mayvjwarpansy15No ratings yet
- Thyroid Disorders in PregnancyDocument46 pagesThyroid Disorders in PregnancyMara Medina - BorleoNo ratings yet
- 2013 Article AbstractsDocument135 pages2013 Article Abstractsmilagros villanuevaNo ratings yet
- Usana Essentials Dubai - Top of The Line, First Class SupplementsDocument2 pagesUsana Essentials Dubai - Top of The Line, First Class SupplementsdxbvvmNo ratings yet
- Age of Exploration Scrapbook: By: Ben SteeleDocument12 pagesAge of Exploration Scrapbook: By: Ben Steeleapi-344972552No ratings yet
- ESP 252 Fang Yuan Bug ListDocument2 pagesESP 252 Fang Yuan Bug ListFang YuanNo ratings yet
- Bolo Vs PerfusiónDocument6 pagesBolo Vs Perfusión1cucu0No ratings yet
- Surgery 3 Questions Exam Template - 0Document8 pagesSurgery 3 Questions Exam Template - 0Morozovschi VitalieNo ratings yet
- Cons Study Summary QuiestionsDocument124 pagesCons Study Summary QuiestionsCons Miyu HimeNo ratings yet
- Nephrotic Syndrome (Nephrosis)Document9 pagesNephrotic Syndrome (Nephrosis)Madhusmita SatapathyNo ratings yet
- Furcation Involvement Classification - A Literature ReviewDocument6 pagesFurcation Involvement Classification - A Literature Reviewsasi dharanNo ratings yet
- CC2D KW Choy - Biochemical Testing in Acute and Chronic Kidney Failure PDFDocument40 pagesCC2D KW Choy - Biochemical Testing in Acute and Chronic Kidney Failure PDFSaad KhanNo ratings yet
- Levy CV Feb 2011Document5 pagesLevy CV Feb 2011jonathan_levy9254No ratings yet
- Answers and RationaleDocument6 pagesAnswers and RationaleSheana TmplNo ratings yet
- ObesityDocument1 pageObesitycaloy10No ratings yet
- Type 1 Diabetes Definitio1Document10 pagesType 1 Diabetes Definitio1Ramya TeddyNo ratings yet