Professional Documents
Culture Documents
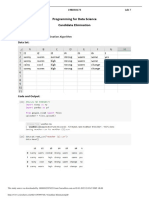
Find S-Algo
Uploaded by
sachin0 ratings0% found this document useful (0 votes)
2 views2 pagesThe document describes several machine learning algorithms:
1) ID3 decision tree algorithm that calculates information gain to determine the best attribute to split on at each node.
2) Naive Bayes algorithm that uses a MultinomialNB classifier to predict text labels after tokenizing and transforming the text into a document-term matrix.
3) Candidate Elimination algorithm that learns concepts by initially setting a specific hypothesis and then generalizing it based on positive and negative examples.
4) Brief mention of an artificial neural network example with input and output arrays.
Original Description:
Original Title
Lab
Copyright
© © All Rights Reserved
Available Formats
PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentThe document describes several machine learning algorithms:
1) ID3 decision tree algorithm that calculates information gain to determine the best attribute to split on at each node.
2) Naive Bayes algorithm that uses a MultinomialNB classifier to predict text labels after tokenizing and transforming the text into a document-term matrix.
3) Candidate Elimination algorithm that learns concepts by initially setting a specific hypothesis and then generalizing it based on positive and negative examples.
4) Brief mention of an artificial neural network example with input and output arrays.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
2 views2 pagesFind S-Algo
Uploaded by
sachinThe document describes several machine learning algorithms:
1) ID3 decision tree algorithm that calculates information gain to determine the best attribute to split on at each node.
2) Naive Bayes algorithm that uses a MultinomialNB classifier to predict text labels after tokenizing and transforming the text into a document-term matrix.
3) Candidate Elimination algorithm that learns concepts by initially setting a specific hypothesis and then generalizing it based on positive and negative examples.
4) Brief mention of an artificial neural network example with input and output arrays.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
You are on page 1of 2
Find S-Algo: def printTree(root: Node, depth=0):
import csv for i in range(depth):
def loadCsv(filename): print("\t", end="")
lines= csv.reader(open(filename, "rt")) print(root.value, end="")
dataset=list(lines) if root.isLeaf:
for i in range(len(dataset)): print("->", root.pred)
dataset[i]=dataset[i] print()
return dataset for child in root.children:
attributes=['Sky', 'Temp', 'Humidity', 'Wind', 'Water', 'Forecast'] printTree(child, depth + 1)
print("\n The attributes are:") root = ID3(data, features)
print(attributes) printTree(root)
num_attributes=len(attributes) NaiveBayes
filename=r"D:\Python\abc.csv" import pandas as pd
dataset=loadCsv(filename) from sklearn.model_selection import train_test_split
print("In The Given Training Data Set:") from sklearn.feature_extraction.text import CountVectorizer
for row in dataset: from sklearn.naive_bayes import MultinomialNB
print(row) from sklearn import metrics
print("\n The target in the Training Data Set:") msg=pd.read_csv('naivetext.csv',names=['message','label'])
target=['Yes', 'Yes', 'No', 'Yes'] print('The dimensions of the dataset',msg.shape)
print(target) msX=msg.message
print("\n The initial value of hypothesis:") y=msg.labelnum
hypothesis=['0']*num_attributes xtrain,xtest,ytrain,ytest=train_test_split(X,y)
print(hypothesis) print ('\n the total number of Training Data :',ytrain.shape)
print("\n Find S: Finding a Maximally Specific Hypothesis") print ('\n the total number of Test Data :',ytest.shape)
for i in range(len(target)): cv = CountVectorizer()
if(target[i]=='Yes'): xtrain_dtm = cv.fit_transform(xtrain)
for j in range(num_attributes): xtest_dtm=cv.transform(xtest)
if(hypothesis[j]=='0'): print('\n The words or Tokens in the text documents \n')
hypothesis[j]=dataset[i][j] print(cv.get_feature_names())
if(hypothesis[j]!=dataset[i][j]): df=pd.DataFrame(xtrain_dtm.toarray(),columns=cv.get_feature_names())
hypothesis[j]='?' clf = MultinomialNB().fit(xtrain_dtm,ytrain)
print(i+1,'=',hypothesis) predicted = clf.predict(xtest_dtm)
print("\n Final Hypothesis") print('\n Accuracy of the classifier is',metrics.accuracy_score(ytest,predicted))
print(hypothesis) print('\n Confusion matrix')
ID3: print(metrics.confusion_matrix(ytest,predicted))
import pandas as pd print('\n The value of Precision', metrics.precision_score(ytest,predicted))
import math print('\nThevalueofRecall',-
import numpy as np metrics.recall_score(ytest,predicted))g['labelnum']=msg.label.map({'pos':1,'neg':0})
data = pd.read_csv("abc.csv") CEA:
features = [feat for feat in data] import numpy as np
print(features) import pandas as pd
features.remove("PlayTennis") data = pd.read_csv('Candidate Elimination Algorithm.csv')
concepts = np.array(data.iloc[:,0:-1])
class Node:
print("\nInstances are:\n",concepts)
def __init__(self): target = np.array(data.iloc[:,-1])
self.children = [] print("\nTarget Values are: ",target)
self.value = "" def learn(concepts, target):
self.isLeaf = False specific_h = concepts[0].copy()
self.pred="" print("\nInitialization of specific_h and genearal_h")
def entropy(examples): print("\nSpecific Boundary: ", specific_h)
general_h = [["?" for i in range(len(specific_h))] for i in range(len(specific_h))]
pos = 0.0
print("\nGeneric Boundary: ",general_h)
neg = 0.0 for i, h in enumerate(concepts):
for _, row in examples.iterrows(): print("\nInstance", i+1 , "is ", h)
if row["PlayTennis"] == "yes": if target[i] == "yes":
pos += 1 print("Instance is Positive ")
else: for x in range(len(specific_h)):
neg += 1 if h[x]!= specific_h[x]:
specific_h[x] ='?'
if pos== 0.0 or neg == 0.0: general_h[x][x] ='?'
return 0.0 if target[i] == "no":
else: print("Instance is Negative ")
p = pos/ (pos + neg) for x in range(len(specific_h)):
n = neg/ (pos+ neg) if h[x]!= specific_h[x]:
return -(p* math.log(p, 2) + n * math.log(n, 2)) general_h[x][x] = specific_h[x]
def info_gain(examples, attr): else:
uniq= np.unique(examples[attr]) general_h[x][x] = '?'
gain = entropy(examples) print("Specific Bundary after ", i+1, "Instance is ", specific_h)
for u in uniq: print("Generic Boundary after ", i+1, "Instance is ", general_h)
subdata = examples[examples[attr] == u] print("\n")
sub_e = entropy(subdata)
gain -= (float(len(subdata)) / float(len(examples))) * sub_e indices = [i for i, val in enumerate(general_h) if val == ['?', '?', '?', '?', '?', '?']]
return gain
def ID3(examples, attrs): for i in indices:
general_h.remove(['?', '?', '?', '?', '?', '?'])
root = Node()
return specific_h, general_h
max_gain = 0
max_feat="" s_final, g_final = learn(concepts, target)
for feature in attrs: print("Final Specific_h: ", s_final, sep="\n")
gain = info_gain(examples, feature) print("Final General_h: ", g_final, sep="\n")
if gain > max_gain:
max_gain = gain ANN:
max_feat = feature import numpy as np
root.value = max_feat X = np.array(([2, 9], [1, 5], [3, 6]), dtype=float)
uniq = np.unique(examples[max_feat]) y = np.array(([92], [86], [89]), dtype=float)
for u in uniq: X = X/np.amax(X,axis=0)
subdata = examples[examples[max_feat] == u] y = y/100
if entropy(subdata) == 0.0: def sigmoid (x):
newNode = Node() return 1/(1 + np.exp(-x))
newNode.isLeaf = True def derivatives_sigmoid(x):
newNode.value = u return x * (1 - x)
newNode.pred = np.unique(subdata["PlayTennis"]) epoch=5
root.children.append(newNode) lr=0.1
else: inputlayer_neurons = 2
dummyNode = Node() hiddenlayer_neurons = 3
dummyNode.value = u output_neurons = 1
new_attrs = attrs.copy() wh=np.random.uniform(size=(inputlayer_neurons,hiddenlayer_neurons))
new_attrs.remove(max_feat) bh=np.random.uniform(size=(1,hiddenlayer_neurons))
child = ID3(subdata, new_attrs) wout=np.random.uniform(size=(hiddenlayer_neurons,output_neurons))
dummyNode.children.append(child) bout=np.random.uniform(size=(1,output_neurons))
root.children.append(dummyNode) for i in range(epoch):
return root hinp1=np.dot(X,wh)
hinp=hinp1 + bh K-Means
hlayer_act = sigmoid(hinp) from sklearn.datasets import load_iris
outinp1=np.dot(hlayer_act,wout) from sklearn.neighbors import KNeighborsClassifier
outinp= outinp1+bout import numpy as np
output = sigmoid(outinp) from sklearn.model_selection import train_test_split
EO = y-output iris_dataset=load_iris()
outgrad = derivatives_sigmoid(output) print("\n IRIS FEATURES \ TARGET NAMES: \n ", iris_dataset.target_names)
d_output = EO * outgrad for i in range(len(iris_dataset.target_names)):
EH = d_output.dot(wout.T) print("\n[{0}]:[{1}]".format(i,iris_dataset.target_names[i]))
hiddengrad = derivatives_sigmoid(hlayer_act) X_train, X_test, y_train, y_test =
d_hiddenlayer = EH * hiddengrad train_test_split(iris_dataset["data"], iris_dataset["target"], random_state=0)
wout += hlayer_act.T.dot(d_output) *lr kn = KNeighborsClassifier(n_neighbors=1)
wh += X.T.dot(d_hiddenlayer) *lr kn.fit(X_train, y_train)
print ("-----------Epoch-", i+1, "Starts----------") x_new = np.array([[5, 2.9, 1, 0.2]])
print("Input: \n" + str(X)) print("\n XNEW \n",x_new)
print("Actual Output: \n" + str(y)) prediction = kn.predict(x_new)
print("Predicted Output: \n" ,output) print("\n Predicted target value: {}\n".format(prediction))
print ("-----------Epoch-", i+1, "Ends----------\n") print("\n Predicted feature name: {}\n".format
print("Input: \n" + str(X)) (iris_dataset["target_names"][prediction]))
print("Actual Output: \n" + str(y)) X_new = np.array([x])
print("Predicted Output: \n" ,output) prediction = kn.predict(X_new)
print("\nActual:{0}{1},Predicted:{2}{3}".format(y_test[i],
Hypothesis Testing: iris_dataset["target_names"][y_test[i]],prediction,iris_dataset["target_names"][prediction]))
import pandas as pd print("\n TEST SCORE[ACCURACY]: {:.2f}\n".format(kn.score(X_test, y_test)))
from scipy import stats
from statsmodels.stats import weightstats as stests LOCALLY WEIGHTED REGRESSION ALGORITHM:
from pprint import pprint import numpy as np
import statsmodels.api as sm import pandas as pd
from statsmodels.formula.api import ols import matplotlib.pyplot as plt
df=pd.read_csv("bp.csv") def kernel (point,xmat, k):
pprint(df[['bp_before','bp_after']].describe()) m,n = np.shape(xmat)
pprint(df.head(5)) weights = np.mat(np.eye((m)))
ttest,pval = stats.ttest_rel(df['bp_before'], df['bp_after']) for j in range(m):
print(pval) diff = point - X[j]
if pval<0.05: weights[j,j] = np.exp(diff*diff.T/(-2.0*k**2))
print("reject null hypothesis") return weights
else: def localweight(point,xmat,ymat,k):
print("accept null hypothesis") wei = kernel(point,xmat,k)
ztest ,pval1 = stests.ztest(df['bp_before'], x2=df['bp_after'], W = (X.T*(wei*X)).I*(X.T*(wei*ymat.T))
value=0,alternative='two-sided') return W
print(float(pval1)) def localweightRegression(xmat,ymat,k):
df_anova = pd.read_csv('PlantGrowth.csv') m,n = np.shape(xmat)
df_anova = df_anova[['weight','group']] ypred = np.zeros(m)
grps = pd.unique(df_anova.group.values) for i in range(m):
d_data = {grp:df_anova['weight'][df_anova.group == grp] for grp in grps} ypred[i] = xmat[i]*localweight(xmat[i],xmat,ymat,k)
F, p = stats.f_oneway(d_data['ctrl'], d_data['trt1'], d_data['trt2']) return ypred
print("p-value for significance is: ", p) def graphPlot(X,ypred):
if p<0.05: sortindex = X[:,1].argsort(0)
print("reject null hypothesis") xsort = X[sortindex][:,0]
else: fig = plt.figure()
print("accept null hypothesis") ax = fig.add_subplot(1,1,1)
df_anova2 = pd.read_csv("https://raw.githubusercontent.com ax.scatter(bill,tip,color='green')
/Opensourcefordatascience/Data-sets/master/crop_yield.csv") ax.plot(xsort[:,1],ypred[sortindex],color='red',linewidth=5)
model = ols('Yield ~ C(Fert)*C(Water)', df_anova2).fit() plt.xlabel('Total bill')
print(f"Overall model F({model.df_model: .0f},{model.df_resid: .0f}) plt.ylabel('Tip')
= {model.fvalue: .3f}, p = {model.f_pvalue: .4f}") plt.show();
print(model.summary()) data = pd.read_csv(r"tips.csv")
res = sm.stats.anova_lm(model, typ= 2) bill = np.array(data.total_bill)
pprint(res) tip = np.array(data.tip)
df_chi = pd.read_csv('chi-test.csv') mbill= np.mat(bill)
contingency_table=pd.crosstab(df_chi["Gender"],df_chi["Like Shopping?"]) mtip= np.mat(tip)
print('contingency_table :-\n',contingency_table) m= np.shape(mbill) [1]
Observed_Values = contingency_table.values one = np.mat (np.ones(m))
print("Observed Values :-\n",Observed_Values) X = np.hstack((one.T, mbill.T))
b=stats.chi2_contingency(contingency_table) ypred=localweightRegression (X,mtip,8)
Expected_Values = b[3] graphPlot(X,ypred)
print("Expected Values :-\n",Expected_Values)
no_of_rows=len(contingency_table.iloc[0:2,0])
no_of_columns=len(contingency_table.iloc[0,0:2])
df11=(no_of_rows-1)*(no_of_columns-1)
print("Degree of Freedom:-",df)
alpha = 0.05
from scipy.stats import chi2
chi_square=sum([(o-e)**2./e for o,e in zip(Observed_Values,Expected_Values)])
chi_square_statistic=chi_square[0]+chi_square[1]
print("chi-square statistic:-",chi_square_statistic)
critical_value=chi2.ppf(q=1-alpha,df=df11)
print('critical_value:',critical_value)
p_value=1-chi2.cdf(x=chi_square_statistic,df=df11)
print('p-value:',p_value)
print('Significance level: ',alpha)
print('Degree of Freedom: ',df11)
print('chi-square statistic:',chi_square_statistic)
print('critical_value:',critical_value)
print('p-value:',p_value)
if chi_square_statistic>=critical_value:
print("Reject H0,There is a relationship between 2 categorical variables")
else:
print("Retain H0,There is no relationship between 2 categorical variables")
if p_value<=alpha:
print("Reject H0,There is a relationship between 2 categorical variables")
else:
print("Retain H0,There is no relationship between 2 categorical variables")
You might also like
- File Handling in PythonDocument25 pagesFile Handling in PythonAlok JhaldiyalNo ratings yet
- Tenth Dimension Ebook PDFDocument227 pagesTenth Dimension Ebook PDFshit-_happens100% (5)
- Finite Element AnalysisDocument160 pagesFinite Element AnalysisRichie Richard0% (1)
- Finite Element Programming With MATLABDocument58 pagesFinite Element Programming With MATLABbharathjoda100% (8)
- Maths Introduction Igcse Extended 3 YrsDocument9 pagesMaths Introduction Igcse Extended 3 YrsYenny TigaNo ratings yet
- Machine Learning Lab (17CSL76)Document48 pagesMachine Learning Lab (17CSL76)Ravi ShankarNo ratings yet
- Machine Learning Lab Record: Dr. Sarika HegdeDocument23 pagesMachine Learning Lab Record: Dr. Sarika Hegde4NM18CS142 SACHIN SINGHNo ratings yet
- Name: Suprit Darshan Shrestha Reg - no:19BCE2584: Lab DA1 Machine Learning LabDocument9 pagesName: Suprit Darshan Shrestha Reg - no:19BCE2584: Lab DA1 Machine Learning LabSuprit D. ShresthaNo ratings yet
- Machine Learning ProgramsDocument17 pagesMachine Learning ProgramsRachana MedehalNo ratings yet
- (P) Program AIODocument22 pages(P) Program AIOYash PraveshNo ratings yet
- ML Lab Programs 1-10-Converted NAM COLLEGE PDFDocument33 pagesML Lab Programs 1-10-Converted NAM COLLEGE PDFPradyumna A KubearNo ratings yet
- Candidate EliminationDocument2 pagesCandidate EliminationSantosh AgadoorappaNo ratings yet
- ML LabDocument7 pagesML LabSharan PatilNo ratings yet
- MLPrograma1-5 PyDocument17 pagesMLPrograma1-5 PybmbzogfxhbufiwsltuNo ratings yet
- Aiml LabDocument14 pagesAiml Lab1DT19IS146TriveniNo ratings yet
- Jntuk R20 MLDocument43 pagesJntuk R20 MLSushNo ratings yet
- Pre-Processing Example - 1Document4 pagesPre-Processing Example - 1Ishani MehtaNo ratings yet
- 11Document10 pages11vachanshetty10No ratings yet
- P 3Document1 pageP 3Fardhin Ahamad ShaikNo ratings yet
- Deep-Learning-Keras-Tensorflow - 1.1.1 Perceptron and Adaline - Ipynb at Master Leriomaggio - Deep-Learning-Keras-TensorflowDocument11 pagesDeep-Learning-Keras-Tensorflow - 1.1.1 Perceptron and Adaline - Ipynb at Master Leriomaggio - Deep-Learning-Keras-Tensorflowme andan buscandoNo ratings yet
- ML Lab Manual PDFDocument9 pagesML Lab Manual PDFAnonymous KvIcYFNo ratings yet
- Import Import DefDocument2 pagesImport Import DefHARSHITHA DNo ratings yet
- ML Lab ManualDocument28 pagesML Lab ManualSharan PatilNo ratings yet
- 15CSL76 StudentsDocument18 pages15CSL76 StudentsYounus KhanNo ratings yet
- 4ann - 5Th ProgDocument15 pages4ann - 5Th Progshreyas .cNo ratings yet
- ML LabDocument7 pagesML LabRishi TPNo ratings yet
- Machine Learning Through Python Lab MannualDocument33 pagesMachine Learning Through Python Lab Mannualjebaf44627No ratings yet
- Machine Learning Laboratory ManualDocument11 pagesMachine Learning Laboratory ManualaakashNo ratings yet
- 6 Task RBFDocument6 pages6 Task RBFJanmejaya SahooNo ratings yet
- Aiml LabDocument13 pagesAiml LabAishwarya BiradarNo ratings yet
- ML RecordDocument18 pagesML Recordharshitsr1234No ratings yet
- ML Lab ManualDocument37 pagesML Lab Manualapekshapandekar01100% (1)
- ML Lab ManualDocument14 pagesML Lab ManualPriya NeelamNo ratings yet
- Ai Exp 4Document9 pagesAi Exp 4aditya bNo ratings yet
- Import Numpy As NPDocument4 pagesImport Numpy As NP4AL19CS059 BHAGYASREENo ratings yet
- ML1Document1 pageML1Afreena afriNo ratings yet
- 'Candidate2.csv': - . - . - in in in in ! in ! in inDocument1 page'Candidate2.csv': - . - . - in in in in ! in ! in inHARSHITHA DNo ratings yet
- المستندDocument2 pagesالمستندهــبه قـاسمNo ratings yet
- SML - Week 2Document4 pagesSML - Week 2szho68No ratings yet
- AstarDocument9 pagesAstaraditya bNo ratings yet
- 1 - All Python Codes + Neo4j SamplesDocument16 pages1 - All Python Codes + Neo4j SamplesSadaf FarooqNo ratings yet
- Experiment 2Document15 pagesExperiment 2saeed wedyanNo ratings yet
- L7 Candidate Elimination PDFDocument5 pagesL7 Candidate Elimination PDFSAMINA ATTARINo ratings yet
- ML p4Document2 pagesML p4Nathon MineNo ratings yet
- FFNNDocument3 pagesFFNNDhanunjay pNo ratings yet
- ML NEW Final FormatDocument37 pagesML NEW Final FormatTharun KumarNo ratings yet
- Candidate Elimination ProblemDocument2 pagesCandidate Elimination ProblemUdupiSri groupNo ratings yet
- Alm Co-3 PDFDocument16 pagesAlm Co-3 PDFThota DeepNo ratings yet
- DAA RecordDocument15 pagesDAA Recorddharun0704No ratings yet
- ML Shristi FileDocument49 pagesML Shristi FileRAVI PARKASHNo ratings yet
- DL 4Document1 pageDL 421jr1a43d1No ratings yet
- 16BCB0126 VL2018195002535 Pe003Document40 pages16BCB0126 VL2018195002535 Pe003MohitNo ratings yet
- ML - LAB - 7 - Jupyter NotebookDocument7 pagesML - LAB - 7 - Jupyter Notebooksuman100% (1)
- C121 Exp1Document32 pagesC121 Exp1Devanshu MaheshwariNo ratings yet
- Perceptron Logic Gates PGMDocument4 pagesPerceptron Logic Gates PGMsanjanshetty3No ratings yet
- Report Q4Document10 pagesReport Q4GAYATHRI NATESHNo ratings yet
- LabDocument25 pagesLabSyed Moizuddin QuadriNo ratings yet
- Is Lab 7Document7 pagesIs Lab 7Aman BansalNo ratings yet
- Is Lab 7Document7 pagesIs Lab 7Aman BansalNo ratings yet
- AIML Lab ProgramsDocument13 pagesAIML Lab ProgramsNeeraja RajeshNo ratings yet
- Find-S Algorithm: Example Sky Airtemp Humidity Wind Water Forecast Enjoysport 1 2 3 4Document3 pagesFind-S Algorithm: Example Sky Airtemp Humidity Wind Water Forecast Enjoysport 1 2 3 4srymegaNo ratings yet
- Ai Asignmnt 2Document7 pagesAi Asignmnt 2Aayush MehtaNo ratings yet
- 04 OOP KindergartenDocument12 pages04 OOP Kindergartengiordano manciniNo ratings yet
- CN SyllabusDocument1 pageCN SyllabussachinNo ratings yet
- Qwedge UpsiDocument1 pageQwedge UpsisachinNo ratings yet
- Gs NotesDocument45 pagesGs NotessachinNo ratings yet
- Gs 3... 1Document7 pagesGs 3... 1sachinNo ratings yet
- 5135 Rover Ruckus Engineering NotebookDocument164 pages5135 Rover Ruckus Engineering Notebookapi-502004424No ratings yet
- Jalali@mshdiua - Ac.ir Jalali - Mshdiau.ac - Ir: Data MiningDocument50 pagesJalali@mshdiua - Ac.ir Jalali - Mshdiau.ac - Ir: Data MiningMostafa HeidaryNo ratings yet
- A Practical Software Package For Computing GravimeDocument13 pagesA Practical Software Package For Computing GravimeandenetNo ratings yet
- Excel DataDocument50 pagesExcel DataninthsevenNo ratings yet
- Operational Objectives DefinitionDocument1 pageOperational Objectives DefinitionZven BlackNo ratings yet
- CBSE Class 12 Chem Notes Question Bank Chemical Kinetics PDFDocument23 pagesCBSE Class 12 Chem Notes Question Bank Chemical Kinetics PDFAshika D ChandavarkarNo ratings yet
- Soal Text GREETING CARDDocument1 pageSoal Text GREETING CARDuswatun hasanahNo ratings yet
- AvizvaDocument2 pagesAvizvaShashi Kant RawatNo ratings yet
- 05 Modeling Dynamic and Static Behavior of Chemical ProcessesDocument47 pages05 Modeling Dynamic and Static Behavior of Chemical ProcessesAsrul SaniNo ratings yet
- Week 4 Day 4 (Lesson 11-Subtask 1)Document7 pagesWeek 4 Day 4 (Lesson 11-Subtask 1)PatzAlzateParaguyaNo ratings yet
- T 3 S 3Document7 pagesT 3 S 3Ashim KumarNo ratings yet
- Taleb - Against VaRDocument4 pagesTaleb - Against VaRShyamal VermaNo ratings yet
- Bahan CR 2Document19 pagesBahan CR 2Exty RikaNo ratings yet
- BroombastickDocument3 pagesBroombastickAllen SornitNo ratings yet
- 2020 MMP-AI Assignment 01 PDFDocument2 pages2020 MMP-AI Assignment 01 PDFpalakNo ratings yet
- NEOM Final RevDocument15 pagesNEOM Final RevSanyam JainNo ratings yet
- Introduction To Systems Approach in Water Quality Decision ProblemsDocument3 pagesIntroduction To Systems Approach in Water Quality Decision ProblemsMALL SUBNo ratings yet
- Lead4ward IQ ToolDocument14 pagesLead4ward IQ TooldowntownameerNo ratings yet
- Stata ExcelDocument44 pagesStata ExcelchompoonootNo ratings yet
- SB PDFDocument197 pagesSB PDFPrabhat SinghNo ratings yet
- Introduction Robotics Lecture1 PDFDocument41 pagesIntroduction Robotics Lecture1 PDFrajavel777No ratings yet
- Divison Unified Exam Oralcomm 1920Document5 pagesDivison Unified Exam Oralcomm 1920William BalalaNo ratings yet
- Electric Field Mapping: Experiment 1Document5 pagesElectric Field Mapping: Experiment 1Pi PoliNo ratings yet
- Precalculus ch6 ReviewDocument2 pagesPrecalculus ch6 Reviewapi-213604106No ratings yet
- Aluminium Weld Fatigue PDFDocument22 pagesAluminium Weld Fatigue PDFMyozin HtutNo ratings yet
- 15 Famous Greek Mathematicians and Their ContributionsDocument16 pages15 Famous Greek Mathematicians and Their ContributionsMallari MarjorieNo ratings yet