Professional Documents
Culture Documents
Unpaired Diseased
Uploaded by
Ionut Sorin0 ratings0% found this document useful (0 votes)
5 views2 pagesThe document analyzes test result data from two medical tests to calculate their sensitivities and compare their performance. It loads the raw data, defines cross tables to calculate test sensitivities. A chi-squared test and Fisher's exact test are performed, finding a statistically significant difference between the two tests with a p-value of 0.026 and odds ratio of 1.95 favoring Test 1.
Original Description:
Copyright
© © All Rights Reserved
Available Formats
TXT, PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentThe document analyzes test result data from two medical tests to calculate their sensitivities and compare their performance. It loads the raw data, defines cross tables to calculate test sensitivities. A chi-squared test and Fisher's exact test are performed, finding a statistically significant difference between the two tests with a p-value of 0.026 and odds ratio of 1.95 favoring Test 1.
Copyright:
© All Rights Reserved
Available Formats
Download as TXT, PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
5 views2 pagesUnpaired Diseased
Uploaded by
Ionut SorinThe document analyzes test result data from two medical tests to calculate their sensitivities and compare their performance. It loads the raw data, defines cross tables to calculate test sensitivities. A chi-squared test and Fisher's exact test are performed, finding a statistically significant difference between the two tests with a p-value of 0.026 and odds ratio of 1.95 favoring Test 1.
Copyright:
© All Rights Reserved
Available Formats
Download as TXT, PDF, TXT or read online from Scribd
You are on page 1of 2
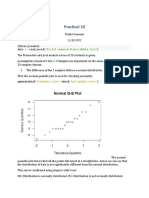
## Get in the data ##
unpaired <- read.csv("unpaired_diseased.csv")
unpaired
test result count
1 0 0 82
2 0 1 18
3 1 0 140
4 1 1 60
## Define the crosstab ##
## Define the counts ##
unpaired_diseased <- tapply(unpaired$count,list(unpaired$test,unpaired$result),c)
## Define labels ##
dimnames(unpaired_diseased) <- list(c("Test 1","Test 2"),c("Result
positive","Result negative"))
unpaired_diseased
Result positive Result negative
Test 1 82 18
Test 2 140 60
## Calculate sensitivities for the two tests ##
library(gmodels)
unpaired_diseased_prop <- CrossTable(unpaired_diseased)
## Display the calculated sensitivities ##
unpaired_diseased_prop$prop.row
Result positive Result negative
Test 1 0.82 0.18
Test 2 0.70 0.30
## Load library for Chi-squared test / Fisher's exact test ##
library(exact2x2)
## Chi-squared Test / Fisher's Exact Test ##
fisher.test(unpaired_diseased)
Fisher's Exact Test for Count Data
data: unpaired_diseased
p-value = 0.02617
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
1.048452 3.757027
sample estimates:
odds ratio
1.948250
You might also like
- Sample Size for Analytical Surveys, Using a Pretest-Posttest-Comparison-Group DesignFrom EverandSample Size for Analytical Surveys, Using a Pretest-Posttest-Comparison-Group DesignNo ratings yet
- Bioestadistica: Clara Carner 2023-05-29Document4 pagesBioestadistica: Clara Carner 2023-05-29Clara CarnerNo ratings yet
- 20BCE1205 Lab6Document12 pages20BCE1205 Lab6SHUBHAM OJHANo ratings yet
- Ancova in R HandoutDocument8 pagesAncova in R HandoutEduardoNo ratings yet
- Software Testing File LovishDocument30 pagesSoftware Testing File LovishMohit SinglaNo ratings yet
- SCRIPT & CONSOLE Non Parametric Summary LarasDocument10 pagesSCRIPT & CONSOLE Non Parametric Summary LarasJessy SeptalistaNo ratings yet
- Stat 2Document3 pagesStat 2smrutiranjan paridaNo ratings yet
- Caso High School and SurveyDocument22 pagesCaso High School and SurveyAntonio ReyesNo ratings yet
- 3 Hypothesis Testing For MeansDocument23 pages3 Hypothesis Testing For Meansmadrazo1423No ratings yet
- Test For The Mean. Population Variance KnownDocument2 pagesTest For The Mean. Population Variance KnownBanti TravelNo ratings yet
- HW 3Document9 pagesHW 3Andalib ShamsNo ratings yet
- Name: Reg. No.: Lab Exercise:: Shivam Batra 19BPS1131Document10 pagesName: Reg. No.: Lab Exercise:: Shivam Batra 19BPS1131Shivam Batra100% (1)
- Week 2Document7 pagesWeek 2Indri Br SitumorangNo ratings yet
- Non Parametric Homework-9Document6 pagesNon Parametric Homework-9Arjun BhusalNo ratings yet
- Lab 6 - ShellDocument7 pagesLab 6 - ShellMansiNo ratings yet
- Regression Analysis ScriptDocument24 pagesRegression Analysis ScriptJohn Riel CaneteNo ratings yet
- Bs at HSW: Victor Omondi 7/27/2021 #Stem Majors ChisquareDocument12 pagesBs at HSW: Victor Omondi 7/27/2021 #Stem Majors ChisquareomondivkeyNo ratings yet
- Comparative Study of ML Algorithms For Heart Disease Prediction Paper Journal IjtrsDocument6 pagesComparative Study of ML Algorithms For Heart Disease Prediction Paper Journal IjtrsSanskar AggarwalNo ratings yet
- StaticsTestDocument6 pagesStaticsTestSOWMIA R 2248056No ratings yet
- ScreenDocument2 pagesScreenIonut SorinNo ratings yet
- Prac 7 Hypothesis TestingDocument1 pagePrac 7 Hypothesis Testingdummy10802No ratings yet
- Lecture7 by Tony - KooDocument22 pagesLecture7 by Tony - Kooclaire.zqmNo ratings yet
- CSC103-Programming Fundamentals: Selections Lecture-8Document35 pagesCSC103-Programming Fundamentals: Selections Lecture-8Gul khanNo ratings yet
- User-Sem-Lavaan (SEM)Document33 pagesUser-Sem-Lavaan (SEM)Meylin ChatherineNo ratings yet
- K Nearest Neighbours (KNN) : Short Intro To KNNDocument13 pagesK Nearest Neighbours (KNN) : Short Intro To KNNLuka FilipovicNo ratings yet
- Sestrada Logistic Regression in R 02172023Document25 pagesSestrada Logistic Regression in R 02172023Mehdi Ben HammiNo ratings yet
- System Verilog RandomizationDocument97 pagesSystem Verilog RandomizationMeghana VeggalamNo ratings yet
- Data Science Comp 2Document13 pagesData Science Comp 2tobiasNo ratings yet
- Practical-10Document4 pagesPractical-10yoshitaNo ratings yet
- R Code Default Data PDFDocument10 pagesR Code Default Data PDFShubham Wadhwa 23No ratings yet
- SMM Lab Ex 1 A) B) C)Document3 pagesSMM Lab Ex 1 A) B) C)prakaash A SNo ratings yet
- 18sets 1Document20 pages18sets 1Dan JoshuaNo ratings yet
- Randomization in SystemVerilogDocument30 pagesRandomization in SystemVerilogSudeep KrishnappaNo ratings yet
- R Notes For Data Analysis and Statistical InferenceDocument10 pagesR Notes For Data Analysis and Statistical InferencergardnercookNo ratings yet
- Uji-Proporsi-dan-Goodness-of-Fit-Test SukmawatiDocument2 pagesUji-Proporsi-dan-Goodness-of-Fit-Test Sukmawatiskmaskm21No ratings yet
- Frequencies: Frequencies Variables Umur /statistics Minimum Maximum /order AnalysisDocument6 pagesFrequencies: Frequencies Variables Umur /statistics Minimum Maximum /order AnalysisInka Rindy AntikaNo ratings yet
- Statistical Analysis Using SASDocument47 pagesStatistical Analysis Using SASMuneesh BajpaiNo ratings yet
- Taller 10Document4 pagesTaller 10sanper.rzvNo ratings yet
- Practical Machine LearningDocument11 pagesPractical Machine Learningminhajur rahmanNo ratings yet
- R PracticeDocument38 pagesR PracticeyoshitaNo ratings yet
- Statistical Computing by Using RDocument11 pagesStatistical Computing by Using RChen-Pan Liao100% (1)
- STA1007S Lab 10: Confidence Intervals: October 2020Document5 pagesSTA1007S Lab 10: Confidence Intervals: October 2020mlunguNo ratings yet
- Linear-Congruential-Method: 1 Avaliação 1: Método Congruencial Linear (MCL)Document12 pagesLinear-Congruential-Method: 1 Avaliação 1: Método Congruencial Linear (MCL)Renato MarquesNo ratings yet
- One Sample Test of HypothesisDocument20 pagesOne Sample Test of HypothesissdfabiaaNo ratings yet
- Learn Python 3 - Python - Code Challenges (Optional) Cheatsheet - CodecademyDocument12 pagesLearn Python 3 - Python - Code Challenges (Optional) Cheatsheet - CodecademyRiaz KhanNo ratings yet
- Maths Lab Assignment 3dfDocument15 pagesMaths Lab Assignment 3dfShreyansh TripathiNo ratings yet
- Reliability: NotesDocument6 pagesReliability: NotesIsmi UnicaNo ratings yet
- Shark Tank Deal Prediction - Uudhya - Dec 2019Document16 pagesShark Tank Deal Prediction - Uudhya - Dec 2019usoramaNo ratings yet
- Maths Lab Assignment 3Document15 pagesMaths Lab Assignment 3Shreyansh TripathiNo ratings yet
- Lab ManualDocument35 pagesLab ManualPraveen KumarNo ratings yet
- ExceptionDocument1 pageExceptionMajjiga KartheekNo ratings yet
- CH4304EXPTSDocument30 pagesCH4304EXPTSAhren PageNo ratings yet
- Trimmed Sample Means For Uniform Mean Estimation and RegressionDocument36 pagesTrimmed Sample Means For Uniform Mean Estimation and RegressionrimfoNo ratings yet
- Multiple Linear RegressionDocument14 pagesMultiple Linear RegressionNamdev100% (1)
- Genetic AlgorithmDocument6 pagesGenetic AlgorithmFilip MajstorovićNo ratings yet
- XSTK Câu hỏiDocument19 pagesXSTK Câu hỏiimaboneofmyswordNo ratings yet
- RA - Assg1: 0.1 AssignmentDocument6 pagesRA - Assg1: 0.1 AssignmentNabhan Abdulla pvNo ratings yet
- Question No1Document6 pagesQuestion No1aniketgupta8433244176No ratings yet
- Lab 5 - ShellDocument7 pagesLab 5 - ShellMansiNo ratings yet
- A1Document8 pagesA1DASHPAGALNo ratings yet
- RocDocument1 pageRocIonut SorinNo ratings yet
- Paired NogoldDocument1 pagePaired NogoldIonut SorinNo ratings yet
- ScreenDocument2 pagesScreenIonut SorinNo ratings yet
- PancreaticDocument2 pagesPancreaticIonut SorinNo ratings yet
- Paired GoldDocument1 pagePaired GoldIonut SorinNo ratings yet