Professional Documents
Culture Documents
Rahul Test Report
Uploaded by
NikHilPaTil0 ratings0% found this document useful (0 votes)
16 views1 pageCopyright
© © All Rights Reserved
Available Formats
PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
16 views1 pageRahul Test Report
Uploaded by
NikHilPaTilCopyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
You are on page 1of 1
Patient's name : Mr Rahul Shanbag Age/Sex : 31 Years Male
Referred by : Dr. Bhargav N Patel Md(micro) Reg. No : 2241
Date : 25/12/2020 Mobile : 7045931211
Ref No. : COVID-6471-25
Patient's Id : HM51
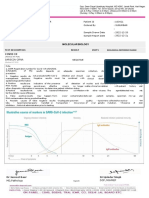
SARS-Cov-2 (COVID - 19) QUALITATIVE RT-PCR
METHOD: Real Time RT-PCR (Qualitative) ICMR Reg No.: CUHPLAG
RdRp gene: TARGET NOT DETECTED
TARGET NOT DETECTED
E gene:
N gene: TARGET NOT DETECTED
Conclusion: COVID – 19 NEGATIVE
Panel Comments:
This molecular test uses Real time PCR technology based on nucleic acid amplification assay for qualitative detection of RNA of Novel
Coronavirus (Covid-19) from Respiratory samples (Throat, Nasopharyngeal swab, BAL fluid & sputum samples). It is an in-vitro diagnostic
test that detects very low levels of COVID-19 RNA in Human clinical samples.
1. "Target Detected" results indicates presence of SARS-Cov-2 in the sample. Positive result does not rule out infection with bacterial or other
viral co-infections.
2. "Target Not Detected" result indicates absence of SARS-Cov-2 infection in the given specimen with the assay used.
A negative result does not exclude the possibility of COVID-19 infection as the results are dependent on many other factors.
Limitations:
1. The results of test are highly dependent on the sampling technique employed, sample type, cold chain maintenance and clinical conditions.
2. FALSE NEGATIVE results may be obtained in case of Presence of PCR inhibitors (Can to be traced by technologists) or viral load lesser
than the assay lower limits of detection as well as rare genotypes or mutations.
3. FALSE POSITIVE may be obtained in case of RNA contamination from preanalytical in lab environment.
4. The assay perfomanance characteristics for this test are determined by ICMR approved manufacturer which is used for clinical diagnosis.
Notes:
1. Results must be interpreted in conjunction with other clinical and/or laboratory findings.
2. Negative results doesn’t not rule out the possibility of COVID-19 infection. Presence of inhibitors in sample, mutations at primer or probe
binding sites or insufficient RNA in patient sample can influence the results.
3. Test reports should be correlated with the clinical presentation and findings.
4. A number of factors could lead to a negative result in an infected individual including
* Poor quality of the specimen, containing inadequate patient material or non-representetive specimen.
* The specimen was collected late or very early in the infection. Optimum specimen types and timing for peak viral levels during infections
caused by 2019-nCoV have not been determined. Collection of multiple samples from the same patient may be necessary to detect the
virus.
* The specimen was not handled and shipped appropriately.
* Technical reasons inherent in the test. e.g Virus mutation or PCR inhibition. * Inadequate numbers of organisms are present in the
specimen.
7. Repeat sampling and testing of lower respiratory specimen is strongly recommended in severe or progressive disease.
8. The repeat specimen ns may be considered after a gap of 2-4 days after the collection of the first specimen for additional testing if required.
9. Categories of viral load is based on cycle threshold (Ct) detected by RT PCR. a.) High viral load : 17 to 24
b.) Moderate viral load : 24 to 31
c.) Low/Mild viral load : 31 to 38
Page 1 of 1
You might also like
- Hepatitis C Virus-Host Interactions and Therapeutics: Current Insights and Future PerspectivesFrom EverandHepatitis C Virus-Host Interactions and Therapeutics: Current Insights and Future PerspectivesNo ratings yet
- Qualitative Detection of Sars-Cov-2 (Covid-19) by RT-PCR: Test ReportDocument1 pageQualitative Detection of Sars-Cov-2 (Covid-19) by RT-PCR: Test Report0001No ratings yet
- Department of Molecular Biology. Covid 19 Test Name Result Unit Bio. Ref. Range MethodDocument2 pagesDepartment of Molecular Biology. Covid 19 Test Name Result Unit Bio. Ref. Range MethodPrantik MaityNo ratings yet
- Irfan Shaikh 38Document2 pagesIrfan Shaikh 38Altamash AnsariNo ratings yet
- Screenshot 2022-04-09 at 10.01.10 AMDocument1 pageScreenshot 2022-04-09 at 10.01.10 AMSachin metkarNo ratings yet
- Department of Molecular Biology. Covid 19 Test Name Result Unit Bio. Ref. Range MethodDocument2 pagesDepartment of Molecular Biology. Covid 19 Test Name Result Unit Bio. Ref. Range MethodManoj NainNo ratings yet
- Sars-Cov2 (Covid-19) Real Time RT PCR TestDocument1 pageSars-Cov2 (Covid-19) Real Time RT PCR Testsanjana rohiteNo ratings yet
- TestReport 203300095Document1 pageTestReport 203300095Sravan KrNo ratings yet
- Mr. Ramkrishan Keshrwani - REPORTDocument1 pageMr. Ramkrishan Keshrwani - REPORTvaibhav vinkareNo ratings yet
- A360086804: Patient ID 136089873 Sid No Cov GNP Branch Mr. Jagathes TDocument1 pageA360086804: Patient ID 136089873 Sid No Cov GNP Branch Mr. Jagathes Tfracncchu CNo ratings yet
- Meera FDocument1 pageMeera FIMOUNT ONENo ratings yet
- Some Tests Are Still in Progress. Report Will Be Available Once All Tests Are CompletedDocument3 pagesSome Tests Are Still in Progress. Report Will Be Available Once All Tests Are CompletedDheeman BaruaNo ratings yet
- Department of Molecular Biology. Covid 19 Test Name Result Unit Bio. Ref. Range MethodDocument2 pagesDepartment of Molecular Biology. Covid 19 Test Name Result Unit Bio. Ref. Range MethodRahul SankaranNo ratings yet
- Covid ReportDocument5 pagesCovid Reportraojip1232No ratings yet
- Mr. Jainil Patel 10 Years: COVID19 Qualitative by Real Time PCRDocument1 pageMr. Jainil Patel 10 Years: COVID19 Qualitative by Real Time PCRIMOUNT ONENo ratings yet
- Covid Report PDFDocument2 pagesCovid Report PDFAthira NairNo ratings yet
- COVID-19 Test Report for Mr. AUDARYA MANEDocument1 pageCOVID-19 Test Report for Mr. AUDARYA MANENeutral GodNo ratings yet
- Sars-Cov2 (Covid-19) Real Time RT PCR TestDocument1 pageSars-Cov2 (Covid-19) Real Time RT PCR TestRoshanNo ratings yet
- COVID-19 PCR Test Report: PositiveDocument1 pageCOVID-19 PCR Test Report: Positiveom agencyNo ratings yet
- Genoamp® Real-Time PCR Tests For Detection of Covid-19: Mahtas Covid-19 Rapid Evidence UpdatesDocument5 pagesGenoamp® Real-Time PCR Tests For Detection of Covid-19: Mahtas Covid-19 Rapid Evidence UpdatesDesmond KhorNo ratings yet
- RTPCRDocument1 pageRTPCRVouch Pro AdminNo ratings yet
- Kavan FDocument1 pageKavan FIMOUNT ONENo ratings yet
- Department of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodDocument2 pagesDepartment of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodAshwini PrinceNo ratings yet
- Mr. Haan Kanuga 3 Years 20100117282: COVID19 Qualitative by Real Time PCRDocument1 pageMr. Haan Kanuga 3 Years 20100117282: COVID19 Qualitative by Real Time PCRIMOUNT ONENo ratings yet
- COVID19 Qualitative by Real Time PCR: COVID19 Interpretation Positive N Gene (CT) 27 Orf Gene (CT) 24 Test: MethodologyDocument1 pageCOVID19 Qualitative by Real Time PCR: COVID19 Interpretation Positive N Gene (CT) 27 Orf Gene (CT) 24 Test: MethodologyNihar DaveNo ratings yet
- Department of Molecular Biology. Covid 19 Test Name Result Unit Bio. Ref. Range MethodDocument4 pagesDepartment of Molecular Biology. Covid 19 Test Name Result Unit Bio. Ref. Range Methodsourabhshrivastava80No ratings yet
- DGRPOPV137Document2 pagesDGRPOPV137Chandni BhaniramkaNo ratings yet
- Department of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodDocument2 pagesDepartment of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodShravan RajavathNo ratings yet
- Qualitative Detection of COVID-19Document1 pageQualitative Detection of COVID-19jogenderNo ratings yet
- Enali FDocument1 pageEnali FIMOUNT ONENo ratings yet
- RTPCR TestDocument1 pageRTPCR TestThe KeyinfraNo ratings yet
- Test Report: (Icmr Registration No.Document1 pageTest Report: (Icmr Registration No.leepisNo ratings yet
- Covid ReportDocument1 pageCovid ReportniketaNo ratings yet
- Department of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodDocument2 pagesDepartment of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodGirija Prasad SwainNo ratings yet
- RTPCR PranavDocument1 pageRTPCR PranavPranav TiwariNo ratings yet
- Od 211570966021947000Document2 pagesOd 211570966021947000Naresh KomaraNo ratings yet
- Report-2210631115831 SHRIYA R 04jan2022 085844Document2 pagesReport-2210631115831 SHRIYA R 04jan2022 085844Shriya RameshNo ratings yet
- Sars-Cov-2 Detection by RT PCR: Req. No: 1121030093Document1 pageSars-Cov-2 Detection by RT PCR: Req. No: 1121030093Hemanth ChowdharyNo ratings yet
- Naidu ReportDocument1 pageNaidu ReportHemanth ChowdharyNo ratings yet
- Sheeba SaleemaDocument1 pageSheeba SaleemaSAMIKSHA GHOSHALNo ratings yet
- Divya Bangera MBBS, MD Microbiology MME Team LeadDocument2 pagesDivya Bangera MBBS, MD Microbiology MME Team LeadRajavardhanNo ratings yet
- 21122557122c Mr. Devki Nandan PunethaDocument2 pages21122557122c Mr. Devki Nandan PunethaDevkinandan PunethaNo ratings yet
- Department of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodDocument2 pagesDepartment of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodadnanpceNo ratings yet
- 0 - Basti Andriyoko, DR., SPPK (K) - Diagnostik Molekuler SARS CoV2. PDS PatKLIn 20102020Document20 pages0 - Basti Andriyoko, DR., SPPK (K) - Diagnostik Molekuler SARS CoV2. PDS PatKLIn 20102020SuliarniNo ratings yet
- Sars-Cov-2 Rna, QL, RT PCR (Covid-19)Document2 pagesSars-Cov-2 Rna, QL, RT PCR (Covid-19)Kathy FuentesNo ratings yet
- Department of Molecular Biology Covid-19 Virus Qualitative PCRDocument2 pagesDepartment of Molecular Biology Covid-19 Virus Qualitative PCRpooja sharmaNo ratings yet
- Report: Covid-19 (Sars-Cov-2) Testing by Real-Time PCRDocument2 pagesReport: Covid-19 (Sars-Cov-2) Testing by Real-Time PCRSidhant DarekarNo ratings yet
- Quality healthcare human right COVID-19 test reportDocument1 pageQuality healthcare human right COVID-19 test reportBhagat SinghNo ratings yet
- Dinesh Poojari0 - ReportDocument1 pageDinesh Poojari0 - ReportRajesh KambleNo ratings yet
- Department of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodDocument3 pagesDepartment of Molecular Biology. Covid 19 RTPCR With Home Collection Test Name Result Unit Bio. Ref. Range MethodPraveen KumarNo ratings yet
- Ms. SUSHILA FUNDE0 - REPORTDocument1 pageMs. SUSHILA FUNDE0 - REPORTSHITAL KHEDKARNo ratings yet
- Testing LabDocument1 pageTesting LabUday TiwariNo ratings yet
- Qualitative Detection of Covid-19 by RT-PCRDocument1 pageQualitative Detection of Covid-19 by RT-PCRRohan DhawaNo ratings yet
- COVID19 Qualitative by Real Time PCRDocument1 pageCOVID19 Qualitative by Real Time PCRNikhil JoseNo ratings yet
- Testing Lab: Quality Healthcare Is A Human RightDocument1 pageTesting Lab: Quality Healthcare Is A Human RightRaghuraj BhatiaNo ratings yet
- ResultDocument1 pageResultNandini Pritesh PatelNo ratings yet
- Covid-19 PCR Test PositiveDocument1 pageCovid-19 PCR Test PositiveBibhas MajumderNo ratings yet
- RTPCR Oct 21Document1 pageRTPCR Oct 21RoshanNo ratings yet
- Noor Mohammad RTPCR Apollo 21012022Document2 pagesNoor Mohammad RTPCR Apollo 21012022DreamNo ratings yet
- Report of Mr. Anand NelsonDocument1 pageReport of Mr. Anand Nelsontejeswini albertNo ratings yet
- GD PI AnthologyDocument541 pagesGD PI AnthologyShailesh KamathNo ratings yet
- GM Diet PlanDocument12 pagesGM Diet PlanNikHilPaTilNo ratings yet
- Brand Revitalisation Strategies for ArrowDocument15 pagesBrand Revitalisation Strategies for ArrowNikHilPaTilNo ratings yet
- Brand Revitalisation Strategies for ArrowDocument15 pagesBrand Revitalisation Strategies for ArrowNikHilPaTilNo ratings yet
- British Petroleum (A) : Defining A Strategic VisionDocument9 pagesBritish Petroleum (A) : Defining A Strategic VisionNikHilPaTilNo ratings yet
- Brand Rahul RedesigningDocument4 pagesBrand Rahul RedesigningNikHilPaTilNo ratings yet
- Brand Revitalisation Strategies for ArrowDocument15 pagesBrand Revitalisation Strategies for ArrowNikHilPaTilNo ratings yet
- Brand Revitalisation Strategies for ArrowDocument15 pagesBrand Revitalisation Strategies for ArrowNikHilPaTilNo ratings yet
- Match Jobs Pictures Sentences Verbs Phrases HobbiesDocument4 pagesMatch Jobs Pictures Sentences Verbs Phrases HobbiesJose ArreagaNo ratings yet
- Surgical Handpiece Maintenance PosterDocument2 pagesSurgical Handpiece Maintenance PosterHayes MaineNo ratings yet
- The Death Penalty-Literature ReviewDocument4 pagesThe Death Penalty-Literature Reviewapi-582834189No ratings yet
- SX SeriesDocument6 pagesSX SeriesmattuttezNo ratings yet
- Census of India 2011 Village and Town Level Data for Purba Champaran District, BiharDocument368 pagesCensus of India 2011 Village and Town Level Data for Purba Champaran District, BiharRahul SharmaNo ratings yet
- Designing a Chlorobenzene PlantDocument13 pagesDesigning a Chlorobenzene PlantAram Nasih MuhammadNo ratings yet
- Condensate Drain Calculation - Lab AHU PDFDocument1 pageCondensate Drain Calculation - Lab AHU PDFAltaf KhanNo ratings yet
- PAC and DELC indicate risk of heart attackDocument3 pagesPAC and DELC indicate risk of heart attackMatheus MoraisNo ratings yet
- WRAP EMS Guide Mar2015Document64 pagesWRAP EMS Guide Mar2015mike24872267No ratings yet
- In Situ Cell Death KitDocument27 pagesIn Situ Cell Death KitckapodisNo ratings yet
- Wisdom Chi KungDocument0 pagesWisdom Chi KungDevlinPyxNo ratings yet
- Geotechnical Stability Guidelines PDFDocument24 pagesGeotechnical Stability Guidelines PDFAhmad Syafiq YudiansyahNo ratings yet
- The Good NurseDocument2 pagesThe Good NurseKiela Therese LabroNo ratings yet
- Dating Questions and AnswersDocument13 pagesDating Questions and AnswersFelicia Babrant98% (49)
- Chapter 3 - HTT547Document33 pagesChapter 3 - HTT547Faadhil MahruzNo ratings yet
- 12 Chemistry Aldehydes Ketones and Carboxylic Acids Test 04 PDFDocument1 page12 Chemistry Aldehydes Ketones and Carboxylic Acids Test 04 PDFShreyash KolekarNo ratings yet
- Covid19 - Attendance Book, Visitors Book, Field AuditDocument7 pagesCovid19 - Attendance Book, Visitors Book, Field AuditmakhalNo ratings yet
- Wire Rope Slings Si 2 - 2 EmmDocument2 pagesWire Rope Slings Si 2 - 2 EmmheppyfaebanffNo ratings yet
- BASF Puristar R3-12 - BF-9220 - PuriStar - R3-12 - PDS - Rev.2020-12 Spec SheetDocument2 pagesBASF Puristar R3-12 - BF-9220 - PuriStar - R3-12 - PDS - Rev.2020-12 Spec SheetAlNo ratings yet
- Standard QAPDocument9 pagesStandard QAPsivaNo ratings yet
- DAPHNE GREASE MP NO.2Document8 pagesDAPHNE GREASE MP NO.2sanusi.pdkmNo ratings yet
- SvagreementDocument28 pagesSvagreementRowena RayosNo ratings yet
- Self-Confidence and Satisfaction Among Nursing Students With The Use of High Fidelity Simulation at Arab American University, PalestineDocument10 pagesSelf-Confidence and Satisfaction Among Nursing Students With The Use of High Fidelity Simulation at Arab American University, PalestineArianna Jasmine MabungaNo ratings yet
- Red Biotechnology ProjectDocument5 pagesRed Biotechnology ProjectMahendrakumar ManiNo ratings yet
- The Bone DreamingDocument3 pagesThe Bone DreamingastrozzNo ratings yet
- Effects of Restorative Materials On Dental PulpDocument32 pagesEffects of Restorative Materials On Dental PulpUpasana BhandariNo ratings yet
- Presentation 1Document20 pagesPresentation 1anon_658550121No ratings yet
- Goat anatomy and physiology guideDocument8 pagesGoat anatomy and physiology guideLochi GmNo ratings yet
- Concept Map (Cells: Sci Bio)Document1 pageConcept Map (Cells: Sci Bio)lu.justina100% (8)
- Imagicle Solutions Available On Cisco DcloudDocument4 pagesImagicle Solutions Available On Cisco Dcloudchindi.comNo ratings yet