Professional Documents
Culture Documents
Chapter 5 Review
Uploaded by
reema cadienteOriginal Description:
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Chapter 5 Review
Uploaded by
reema cadienteCopyright:
Available Formats
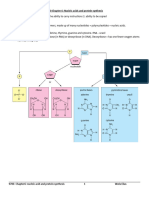
1. What are the structural differences between DNA and RNA? a.
The nitrogenous bases of DNA are Glycine, Cytosine, Thymine and Adenine while the nitrogenous bases of RNA are the same except for Thymine which is replaced with Uracil b. DNA s sugar is deoxyribose, has a hydrogen atom at this position and contains one oxygen atom fewer overall, while RNA s sugar is ribose, sugar that has a hydroxyl group attached to the 2 -carbon atom. c. RNA is single-stranded while DNA is double-stranded with a helical secondary structure 2. Name the enzyme that adds the nucleotides during DNA replication. DNA polymerase III 3. What is the difference between a nucleotide and nucleoside? Nucleotide- consists of a sugar, a phosphate, and a nitrogenous base Nucleoside- a compound consisting of a purine or pyrimidine base linked to a sugar, especially ribose or deoxyribose 4. What are Okazaki fragments? Why are they formed? Short lengths of DNA produced on the lagging strand during DNA replication. They are rapidly joined by DNA ligase to form a continuous DNA strand. They are formed because the DNA polymerase can only polymerize the 5 -3 direction, therefore the lagging strand is first synthesized with RNA primers and then would leave an interval with an open OH for the DNA polymerase to add nucleotides 5. What is meant by semi conservative replication? In a round of replication, each of the two strands of DNA is used as a template for the formation of a complementary DNA strand. The original strands therefore remain intact through many cell generations. 6. List the functions of the DNA polymerase molecule.
7. What enzyme seals the Okazaki fragments? DNA ligase 8. What is a replication fork? The point of unwinding, where the two single nucleotide strands separate from the doublestranded DNA helix 9. What is the advantage of numerous replication forks in replicating DNA molecule? -so that if one fork is damaged, the replication can still proceed in the other fork
-speed 10. Describe the steps involved in DNA replication.
Initiation
Unwinding Elongation Termination
The circular chromosome of E. coli has a single replication origin (oriC). The minimal sequence required for oriC to function consists of 245 bp that contain several critical sites. Initiator proteins bind to oriC and cause a short section of DNA to unwind. This unwinding allows helicase and other single-strand-binding proteins to attach to the polynucleotide strand DNA helicases, SSBP, topoisomerase, primase DNA pol I,II,III, ligase
11. What are the differences in DNA replication between a prokaryote and eukaryote? a. Initiation of replication Prokaryote Replication origins are specified by specific DNA sequences that are only several hundred nucleotides pair long. They also have a single origin of replication in a circular chromosome. Eukaryote The sequences needed to specify an origin of DNA replication seem to be less well-defined, and the origin can span several thousand nucleotide parts. Eukaryotic replication takes places at S phase. The replication fork in eucaryotes moves about t0
times more slowly than the bacterial replication fork, and the much longer eucaryotic chromosomes each require many replication origins to complete their replication in an S phase, which typically lasts for 8 hours in human cells b. Termination of replication Prokaryote Bacteria have a circular DNA as chromosomes in where replication ends when two replication forks meet starting from the initiation point Eukaryote Eukaryotes have specialized nucleotide sequences at the end of the chromosomes that are incorporated into structures called telomeres *telomerase- the enzyme that replenishes the nucleotide sequences every time the cell divides Telomerase extends one of the DNA strands at the end of a chromosome by using an RNA template that is an integral part of the enzyme itself, producing a highly repeated DNA sequence that typically extends for thousands of nucleotide pairs at each chromosome end. c. Lagging strand polymerization Prokaryote 1000-2000 interval Eukaryote About 100-200 nucleotides interval
12. Describe how the following enzymes or proteins would function in DNA replication: a. Primase -synthesizes short RNA primers on the lagging strand which uses a ribonucleoside triphosphates b. Ligase -joins the 3 end of the new DNA fragment to the 5 end of the previous one to produce a continuous DNA chain from the many DNA fragments made on the lagging strand. c. DNA pol 5 -3 polymerizing activity -elongates the DNA d. DNA pol 3 -5 editing function
-3'-to-5 ' exonuclease activity attached to DNA polymerase chews back to create a base paired 3'OH end on the primer strand. DNA polymerase continues the process of adding nucleotides to the base-paired 3-OH end of the primer strand
e. DNA pol 5 -3 exonuclease activity
-Clips off any unpaired residues at the primer terminus, continuing until enough nucleotides have been removed to regenerate a correctly base-paired 3 -OH terminus that can prime DNA synthesis. A self-correcting enzyme that removes its own polymerization errors as it moves along the DNA. f. Initiator protein a) Helps unwind the origin using helicase b) Guides replication machinery to the origin c) Eukaryote: links cell cycle control to origin activation g. Helicase -facilitates the unwinding of the helix to create template strands; accomplished by ATP hydrolysis h. Single strand binding protein -binds tightly and cooperatively to expose s-s DNA w/o covering the bases made available for templating i. Sliding clamp protein - Keeps the polymerase firmly on the DNA when it is moving but releases it as soon as the polymerase runs into a double-stranded DNA Topoisomerase - Add itself covalently to a DNA backbone phosphate thus, breaking a phosphodiester bond in a DNA in a DNA strand
j.
13. Draw a replication fork and label the 5 and 3 ends. Include the leading and lagging strands in your drawing. Beneath the drawing, indicate which direction the replication fork is moving. Why is the 3 OH group on the deoxyribose ring so important for DNA synthesis?
14. What are the three major steps of polymerase chain reaction? a. Denaturation b. Annealing c. Extension/ elongation
You might also like
- The Replication of DNA in Prokaryo1tesDocument7 pagesThe Replication of DNA in Prokaryo1teshgkutjutNo ratings yet
- Chapt 7 DNA Replication NotesDocument8 pagesChapt 7 DNA Replication Notesmalenya1100% (1)
- 637605951536919459classxiibiologych 6PPT 1Document11 pages637605951536919459classxiibiologych 6PPT 1Kamla RathiNo ratings yet
- DNA Replication: The Semi-Conservative ProcessDocument7 pagesDNA Replication: The Semi-Conservative ProcessMuhammad QayumNo ratings yet
- Lecture Notes 8Document5 pagesLecture Notes 8rishabhNo ratings yet
- 14.4 DNA Replication in ProkaryotesDocument3 pages14.4 DNA Replication in ProkaryotesAyushi MauryaNo ratings yet
- Bio001 Balag, MayreisDocument11 pagesBio001 Balag, MayreisMay Reis BalagNo ratings yet
- DNA Replication-Sem 2 20222023-UKMFolio-Part 3Document40 pagesDNA Replication-Sem 2 20222023-UKMFolio-Part 3Keesal SundraNo ratings yet
- 2022-04-17 L4 - DNA ReplicationDocument46 pages2022-04-17 L4 - DNA ReplicationTamara ElyasNo ratings yet
- Dianne D. Mariano BSN 101Document9 pagesDianne D. Mariano BSN 101Dianne MarianoNo ratings yet
- Composed of A BaseDocument23 pagesComposed of A Basetanvi bhallaNo ratings yet
- DNA Synthesis (Replication) - Mechanisms and Types of Replication in Prokaryotes and Eukaryotes.Document11 pagesDNA Synthesis (Replication) - Mechanisms and Types of Replication in Prokaryotes and Eukaryotes.carlottabovi28No ratings yet
- Nucleic acidsLECTUREDocument116 pagesNucleic acidsLECTUREKesha Marie TalloNo ratings yet
- Bio Dna Replication - Transcription and TranslationDocument6 pagesBio Dna Replication - Transcription and TranslationXie YuJiaNo ratings yet
- Conceptual Question Molecular BiologyDocument112 pagesConceptual Question Molecular BiologykoyewubrhanuNo ratings yet
- DNA Replication and Metabolic RegulationDocument3 pagesDNA Replication and Metabolic RegulationDeitherNo ratings yet
- The Central Dogma of Molecular BiologyDocument28 pagesThe Central Dogma of Molecular BiologysannsannNo ratings yet
- DNA ReplicationDocument4 pagesDNA ReplicationJanine San LuisNo ratings yet
- DNA Study GuideDocument4 pagesDNA Study GuidekittihsiangNo ratings yet
- Bacterial GeneticsDocument38 pagesBacterial Geneticsfatima zafarNo ratings yet
- Designer Genes: Practice - Molecular-Genetic GeneticsDocument26 pagesDesigner Genes: Practice - Molecular-Genetic GeneticsGiovanni TorresNo ratings yet
- Genetic Control: By. Yasmine HadiastrianiDocument14 pagesGenetic Control: By. Yasmine HadiastrianiYasmine HadiastrianiNo ratings yet
- 6 BIO462 Chapter 6Document21 pages6 BIO462 Chapter 6Syafiqah SuhaimiNo ratings yet
- Replication and DNA Repair RecitDocument10 pagesReplication and DNA Repair RecithoythereNo ratings yet
- Dna Replication NotesDocument11 pagesDna Replication NotesJanine San LuisNo ratings yet
- 4 DnaDocument6 pages4 Dnadr_47839666No ratings yet
- Eukaryotic_ReplicationDocument11 pagesEukaryotic_ReplicationSuraj DubeyNo ratings yet
- DNA ReplicationDocument18 pagesDNA ReplicationAditi PatriyaNo ratings yet
- DNA ReplicationDocument28 pagesDNA ReplicationAsim Bin Arshad 58-FBAS/BSBIO/S19100% (1)
- DNA Replication NotesDocument2 pagesDNA Replication NotesMart Tin100% (1)
- Nucleic acids structure and functionsDocument56 pagesNucleic acids structure and functionsCr U NchNo ratings yet
- DNA Structure and Replication in Human Population GeneticsDocument3 pagesDNA Structure and Replication in Human Population GeneticsBenjamin Aluoch OnyangoNo ratings yet
- DNA Replication: A Concise GuideDocument39 pagesDNA Replication: A Concise Guidehjklknnm jhoiolkNo ratings yet
- DNA ReplicationDocument47 pagesDNA ReplicationYSOBELLATHERESE BILOLONo ratings yet
- Dna Replication and Repair #4Document76 pagesDna Replication and Repair #4Len ArellanoNo ratings yet
- DNA replication and protein synthesisDocument4 pagesDNA replication and protein synthesisKingNo ratings yet
- BCH 401 Note 1 - KWASUDocument35 pagesBCH 401 Note 1 - KWASUidriscognitoleadsNo ratings yet
- DNA Replication: How Cells Copy Genetic MaterialDocument14 pagesDNA Replication: How Cells Copy Genetic MaterialIlac Tristan BernardoNo ratings yet
- Long Essay 2Document7 pagesLong Essay 2Alexandru-Crivac Cristina-NicoletaNo ratings yet
- DR Linda Molecular BiosDocument73 pagesDR Linda Molecular BiosValency BathoNo ratings yet
- 2 - Molecular Biology and GeneticsDocument14 pages2 - Molecular Biology and Geneticsarahunt253No ratings yet
- (Lecture 2) GenesDocument39 pages(Lecture 2) GenesKasraSrNo ratings yet
- The Biological Information Flows From DNA To RNA and From There To ProteinsDocument38 pagesThe Biological Information Flows From DNA To RNA and From There To Proteinsfentaw melkieNo ratings yet
- DNA ReplicationDocument7 pagesDNA Replicationlubuto TuntepeNo ratings yet
- DNA Replication and RepairDocument7 pagesDNA Replication and RepairJhun Lerry TayanNo ratings yet
- DNA Structure & ReplicationDocument19 pagesDNA Structure & ReplicationShigrid Ann DocilNo ratings yet
- Molecular Biology Study MaterialDocument58 pagesMolecular Biology Study MaterialchitraNo ratings yet
- Packaging of DNA Into ChromosomeDocument13 pagesPackaging of DNA Into ChromosomeShashank AppuNo ratings yet
- DIMA (Slides 1 Through 4)Document2 pagesDIMA (Slides 1 Through 4)Mohamed AlaeddinNo ratings yet
- MODULE 5-GeneticsDocument9 pagesMODULE 5-GeneticsDivine Grace CincoNo ratings yet
- DNA ReplicationDocument16 pagesDNA Replicationwmdpr4x64fNo ratings yet
- Replication Transcription and TranslationDocument16 pagesReplication Transcription and TranslationBalangat Regine L.No ratings yet
- DNA Replication SBI4UDocument6 pagesDNA Replication SBI4UkishoreddiNo ratings yet
- DNA Replication Notes: Purpose: To Make An Identical Copy of DNA For Cell Division Process: BackgroundDocument3 pagesDNA Replication Notes: Purpose: To Make An Identical Copy of DNA For Cell Division Process: BackgroundLuke HughesNo ratings yet
- 56404332dna ReplicationDocument4 pages56404332dna ReplicationKaranNo ratings yet
- Molecular Biology Unit IV and VDocument60 pagesMolecular Biology Unit IV and VchitraNo ratings yet
- 9700 Chapter 6 Nucleic acids and protein synthesis study sheetDocument21 pages9700 Chapter 6 Nucleic acids and protein synthesis study sheetwafa eliasNo ratings yet
- 7 - Nucleic Acids DNA ReplicationDocument4 pages7 - Nucleic Acids DNA ReplicationShardy Lyn RuizNo ratings yet
- Chapter 12 DNA and RNA ReviewDocument6 pagesChapter 12 DNA and RNA ReviewAndrew WatsonNo ratings yet
- Sample Patient HX PDFDocument5 pagesSample Patient HX PDFreema cadienteNo ratings yet
- Holy SpiritDocument1 pageHoly SpiritcrennydaneNo ratings yet
- Why Jose Rizal Is The Greatest National Hero by Elizabeth Ocampo (Book Review)Document5 pagesWhy Jose Rizal Is The Greatest National Hero by Elizabeth Ocampo (Book Review)reema cadiente100% (1)
- EXER2 Fecal Examination For ParasitesDocument6 pagesEXER2 Fecal Examination For Parasitesreema cadienteNo ratings yet
- Exer3 The Protozoans 1Document4 pagesExer3 The Protozoans 1reema cadienteNo ratings yet
- Chinese CivilizationDocument2 pagesChinese Civilizationreema cadienteNo ratings yet
- EXER2 Fecal Examination For ParasitesDocument6 pagesEXER2 Fecal Examination For Parasitesreema cadienteNo ratings yet
- EXER2 Fecal Examination For ParasitesDocument6 pagesEXER2 Fecal Examination For Parasitesreema cadienteNo ratings yet
- Down SyndromeDocument2 pagesDown Syndromereema cadienteNo ratings yet
- Gods Dream by Bo SanchezDocument28 pagesGods Dream by Bo SanchezPauline Joanne Arceo100% (1)
- DNA Replication in Prokaryotes and EukaryotesDocument7 pagesDNA Replication in Prokaryotes and Eukaryotessmn416No ratings yet
- MCB 110 Study GuideDocument10 pagesMCB 110 Study GuideAlexPowersNo ratings yet
- Unit 3Document83 pagesUnit 3ShekharNo ratings yet
- PEGFP-N1 Vector InformationDocument3 pagesPEGFP-N1 Vector InformationNicholas SoNo ratings yet
- Origin of Replication: Gram Negative Bacterial PlasmidsDocument12 pagesOrigin of Replication: Gram Negative Bacterial PlasmidsMagesh RamasamyNo ratings yet
- Eukaryotic DNA Replication OriginsDocument81 pagesEukaryotic DNA Replication OriginsBintang PundhraNo ratings yet
- Session 5Document28 pagesSession 5IndhumathiNo ratings yet
- DNA ReplicationDocument44 pagesDNA ReplicationRana AttiqNo ratings yet
- Genetic Analysis An Integrated Approach 2Nd Edition Sanders Test Bank Full Chapter PDFDocument35 pagesGenetic Analysis An Integrated Approach 2Nd Edition Sanders Test Bank Full Chapter PDFallison.young656100% (15)
- Genetic Analysis An Integrated Approach 2nd Edition Sanders Test Bank 1Document14 pagesGenetic Analysis An Integrated Approach 2nd Edition Sanders Test Bank 1theresa100% (36)
- Rolling Circle PDFDocument20 pagesRolling Circle PDFسٓنج ايكٓNo ratings yet
- pGBKT7 MapDocument0 pagespGBKT7 MapJuan Jose MartinezNo ratings yet
- Bioinformatics Chapter 1Document54 pagesBioinformatics Chapter 1kawchar.husain.85No ratings yet
- L 8 DNA Packaging and ReplicationDocument44 pagesL 8 DNA Packaging and ReplicationsNo ratings yet
- Vector BiologyDocument32 pagesVector BiologymaniiiiiiiiNo ratings yet
- Plasmids 101 Ebook 3rd Ed FinalDocument193 pagesPlasmids 101 Ebook 3rd Ed FinalManuel CamachoNo ratings yet
- Semi-Conservative DNA ReplicationDocument12 pagesSemi-Conservative DNA ReplicationDibyakNo ratings yet
- Microbiology 1st Edition Wessner Test BankDocument18 pagesMicrobiology 1st Edition Wessner Test Bankthomasmayoaecfpmwgn100% (29)
- Pcna, The Maestro of The Replication Fork2007Document15 pagesPcna, The Maestro of The Replication Fork2007gisin touNo ratings yet
- Molecular Biology of The Cell, Sixth Edition Chapter 5: Dna Replication, Repair, and RecombinationDocument25 pagesMolecular Biology of The Cell, Sixth Edition Chapter 5: Dna Replication, Repair, and RecombinationIsmael Torres-PizarroNo ratings yet
- DNA Polymerase IIIDocument24 pagesDNA Polymerase IIIAhmad JavaidNo ratings yet
- Lecture NotesDocument32 pagesLecture Noteslira shresthaNo ratings yet
- Proteins Involved in DNA ReplicationDocument8 pagesProteins Involved in DNA ReplicationThe FourNo ratings yet
- Multiple Replication Forks E.coliDocument18 pagesMultiple Replication Forks E.coliDr. Atif A. PatoliNo ratings yet
- DNA Replication Part I PDFDocument31 pagesDNA Replication Part I PDFAlok PatraNo ratings yet
- Bacteria Eukarya EukaryogenesisDocument20 pagesBacteria Eukarya EukaryogenesisJamesNo ratings yet
- DNA Replication in ProkaryotesDocument28 pagesDNA Replication in ProkaryotesvinodkumarmishraNo ratings yet
- Csir Ugc Net - Life Sciences - Free Sample TheoryDocument12 pagesCsir Ugc Net - Life Sciences - Free Sample TheorySyamala Natarajan100% (1)
- DNA ReplicationDocument28 pagesDNA ReplicationAsim Bin Arshad 58-FBAS/BSBIO/S19100% (1)
- Molecular Biology of The Cell, 5th EditionDocument82 pagesMolecular Biology of The Cell, 5th EditionBee Nunes25% (67)
- Tales from Both Sides of the Brain: A Life in NeuroscienceFrom EverandTales from Both Sides of the Brain: A Life in NeuroscienceRating: 3 out of 5 stars3/5 (18)
- When the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisFrom EverandWhen the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisRating: 3.5 out of 5 stars3.5/5 (2)
- Why We Die: The New Science of Aging and the Quest for ImmortalityFrom EverandWhy We Die: The New Science of Aging and the Quest for ImmortalityRating: 4 out of 5 stars4/5 (3)
- All That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesFrom EverandAll That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesRating: 4.5 out of 5 stars4.5/5 (397)
- A Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsFrom EverandA Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsRating: 4.5 out of 5 stars4.5/5 (4)
- The Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceFrom EverandThe Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceRating: 4.5 out of 5 stars4.5/5 (515)
- 10% Human: How Your Body's Microbes Hold the Key to Health and HappinessFrom Everand10% Human: How Your Body's Microbes Hold the Key to Health and HappinessRating: 4 out of 5 stars4/5 (33)
- Gut: the new and revised Sunday Times bestsellerFrom EverandGut: the new and revised Sunday Times bestsellerRating: 4 out of 5 stars4/5 (392)
- Inside of a Dog: What Dogs See, Smell, and KnowFrom EverandInside of a Dog: What Dogs See, Smell, and KnowRating: 4 out of 5 stars4/5 (390)
- The Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionFrom EverandThe Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionRating: 4 out of 5 stars4/5 (811)
- Gut: The Inside Story of Our Body's Most Underrated Organ (Revised Edition)From EverandGut: The Inside Story of Our Body's Most Underrated Organ (Revised Edition)Rating: 4 out of 5 stars4/5 (378)
- A Series of Fortunate Events: Chance and the Making of the Planet, Life, and YouFrom EverandA Series of Fortunate Events: Chance and the Making of the Planet, Life, and YouRating: 4.5 out of 5 stars4.5/5 (62)
- Wayfinding: The Science and Mystery of How Humans Navigate the WorldFrom EverandWayfinding: The Science and Mystery of How Humans Navigate the WorldRating: 4.5 out of 5 stars4.5/5 (18)
- Who's in Charge?: Free Will and the Science of the BrainFrom EverandWho's in Charge?: Free Will and the Science of the BrainRating: 4 out of 5 stars4/5 (65)
- Moral Tribes: Emotion, Reason, and the Gap Between Us and ThemFrom EverandMoral Tribes: Emotion, Reason, and the Gap Between Us and ThemRating: 4.5 out of 5 stars4.5/5 (115)
- Fast Asleep: Improve Brain Function, Lose Weight, Boost Your Mood, Reduce Stress, and Become a Better SleeperFrom EverandFast Asleep: Improve Brain Function, Lose Weight, Boost Your Mood, Reduce Stress, and Become a Better SleeperRating: 4.5 out of 5 stars4.5/5 (15)
- The Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorFrom EverandThe Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorNo ratings yet
- Undeniable: How Biology Confirms Our Intuition That Life Is DesignedFrom EverandUndeniable: How Biology Confirms Our Intuition That Life Is DesignedRating: 4 out of 5 stars4/5 (11)
- Buddha's Brain: The Practical Neuroscience of Happiness, Love & WisdomFrom EverandBuddha's Brain: The Practical Neuroscience of Happiness, Love & WisdomRating: 4 out of 5 stars4/5 (215)
- Crypt: Life, Death and Disease in the Middle Ages and BeyondFrom EverandCrypt: Life, Death and Disease in the Middle Ages and BeyondRating: 4 out of 5 stars4/5 (4)
- Human: The Science Behind What Makes Your Brain UniqueFrom EverandHuman: The Science Behind What Makes Your Brain UniqueRating: 3.5 out of 5 stars3.5/5 (38)
- The Lives of Bees: The Untold Story of the Honey Bee in the WildFrom EverandThe Lives of Bees: The Untold Story of the Honey Bee in the WildRating: 4.5 out of 5 stars4.5/5 (44)
- Good Without God: What a Billion Nonreligious People Do BelieveFrom EverandGood Without God: What a Billion Nonreligious People Do BelieveRating: 4 out of 5 stars4/5 (66)
- The Dog Who Couldn't Stop Loving: How Dogs Have Captured Our Hearts for Thousands of YearsFrom EverandThe Dog Who Couldn't Stop Loving: How Dogs Have Captured Our Hearts for Thousands of YearsNo ratings yet