Professional Documents
Culture Documents
20BCE2126 ML Da 5
Uploaded by
vedantmodi2020130 ratings0% found this document useful (0 votes)
5 views3 pagesThe document describes an assignment to perform k-means clustering on wine data to group wines based on their malic acid and proline levels, including preprocessing the data, computing the elbow plot to determine the optimal number of clusters k, running k-means clustering, and assigning cluster labels to each data point. The analysis finds an optimal k of 3 clusters for grouping the wines.
Original Description:
Copyright
© © All Rights Reserved
Available Formats
PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentThe document describes an assignment to perform k-means clustering on wine data to group wines based on their malic acid and proline levels, including preprocessing the data, computing the elbow plot to determine the optimal number of clusters k, running k-means clustering, and assigning cluster labels to each data point. The analysis finds an optimal k of 3 clusters for grouping the wines.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
5 views3 pages20BCE2126 ML Da 5
Uploaded by
vedantmodi202013The document describes an assignment to perform k-means clustering on wine data to group wines based on their malic acid and proline levels, including preprocessing the data, computing the elbow plot to determine the optimal number of clusters k, running k-means clustering, and assigning cluster labels to each data point. The analysis finds an optimal k of 3 clusters for grouping the wines.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
You are on page 1of 3
Assignment – 5
Name – Vedant Modi
Reg. no.- 20BCE2126
Course – Machine Learning
Lab – L21+L22
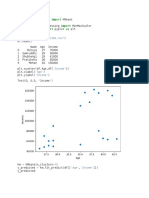
Question : Use wine data set from sklearn library and try to form
clusters of wine behaviour using Malic Acid and Proline features.
Drop the other features for simplicity. -Create a scatter plot of the
above mentioned features of the wine data set. -Figure out if any
pre-processing such as scaling would here. -Draw elbow plot and
from that figure out an optimal value of k.
import pandas as pd
from sklearn.datasets import load_wine
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import KMeans
from matplotlib import pyplot as plt
# Load the wine dataset
wine = load_wine()
# Create a DataFrame with only the Malic Acid and Proline features
data = pd.DataFrame(wine.data[:, [0, 11]], columns=['Malic acid', 'Proline'])
# Create a scatter plot of the Malic Acid and Proline features
plt.scatter(data['Malic acid'], data['Proline'])
plt.xlabel('Malic acid')
plt.ylabel('Proline')
plt.show()
# Standardize the data
scaler = StandardScaler()
scaled_data = scaler.fit_transform(data)
# Compute the sum of squared distances for a range of k values
k_values = range(1, 11)
sse = []
for k in k_values:
kmeans = KMeans(n_clusters=k)
kmeans.fit(scaled_data)
sse.append(kmeans.inertia_)
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will chan
warnings.warn(
# Plot the elbow method
plt.plot(k_values, sse)
plt.xlabel('Number of clusters (k)')
plt.ylabel('Sum of squared distances')
plt.show()
# Determine the optimal number of clusters using the elbow method
optimal_k = 3
# Perform clustering with the optimal number of clusters
kmeans = KMeans(n_clusters=optimal_k)
kmeans.fit(scaled_data)
/usr/local/lib/python3.10/dist-packages/sklearn/cluster/_kmeans.py:870: FutureWar
warnings.warn(
▾ KMeans
KMeans(n_clusters=3)
# Assign cluster labels to each data point
data['cluster'] = kmeans.labels_
# Print the results
print('Cluster labels:')
print(data['cluster'].value_counts())
Cluster labels:
0 65
2 57
1 56
Name: cluster, dtype: int64
You might also like
- Scikit Learn Cheat SheetDocument9 pagesScikit Learn Cheat Sheetburhan ökNo ratings yet
- K MeansDocument329 pagesK Meansyousef shaban100% (1)
- Cluster AlgorithmDocument25 pagesCluster AlgorithmDoli PaikNo ratings yet
- RA2111003011432Document3 pagesRA2111003011432IMMANUEL RUSSO (RA2111051010032)No ratings yet
- KMeansDocument1 pageKMeansbouazizchahine7No ratings yet
- Vid 4Document6 pagesVid 4diyalap01No ratings yet
- KmeansDocument7 pagesKmeanspatil samrudhiNo ratings yet
- R Lab ProgramDocument20 pagesR Lab ProgramRadhiyadevi ChinnasamyNo ratings yet
- Everything Useful I Know About Kubectl - Atomic CommitsDocument10 pagesEverything Useful I Know About Kubectl - Atomic CommitstutorificNo ratings yet
- Output2Document2 pagesOutput2Laptop-Dimas-249No ratings yet
- IRIS Commands PracticeDocument10 pagesIRIS Commands Practiceaqib ahmedNo ratings yet
- Seminar 10Document3 pagesSeminar 10Nishad AhamedNo ratings yet
- ML0101EN Clus DBSCN Weather Py v1Document16 pagesML0101EN Clus DBSCN Weather Py v1Rajat SolankiNo ratings yet
- Deep Learning RecordDocument70 pagesDeep Learning Recordmathan.balaji.02No ratings yet
- Installing Spark On Windows EnvironmentDocument16 pagesInstalling Spark On Windows EnvironmentDr Mohammed KamalNo ratings yet
- ML With Python PracticalDocument22 pagesML With Python Practicaln58648017No ratings yet
- Subject: ML Name: Priyanshu Gandhi Date: 10/4/21 Expt. No.: 9 Roll No.: C008 Title: Clustering Implementation in PythonDocument7 pagesSubject: ML Name: Priyanshu Gandhi Date: 10/4/21 Expt. No.: 9 Roll No.: C008 Title: Clustering Implementation in PythonKartik KatekarNo ratings yet
- ProjectDocument17 pagesProjectmohamed mohsenNo ratings yet
- Ilovepdf MergedDocument25 pagesIlovepdf Mergedrohitfc3105No ratings yet
- K-Means in Python - SolutionDocument6 pagesK-Means in Python - SolutionRodrigo ViolanteNo ratings yet
- Import Pandas As PD DF PD - Read - CSV ("Titanic - Train - CSV") DF - HeadDocument20 pagesImport Pandas As PD DF PD - Read - CSV ("Titanic - Train - CSV") DF - HeadSaloni TuliNo ratings yet
- Assignment 2.1.2 Image AugmentationDocument8 pagesAssignment 2.1.2 Image AugmentationHockhin OoiNo ratings yet
- DeepLearningForVisionSystems Ch5 AlexNetDocument32 pagesDeepLearningForVisionSystems Ch5 AlexNetmkkadambiNo ratings yet
- 2324 BigData Lab3Document6 pages2324 BigData Lab3Elie Al HowayekNo ratings yet
- Data Mining and Warehousing Concepts Lab: (ITPC - 228)Document6 pagesData Mining and Warehousing Concepts Lab: (ITPC - 228)Angelina TutuNo ratings yet
- ML - Practical FileDocument15 pagesML - Practical FileJatin MathurNo ratings yet
- Problem Set 1: Introduction To R - Solutions With R Output: 1 Install PackagesDocument24 pagesProblem Set 1: Introduction To R - Solutions With R Output: 1 Install PackagesDarnell LarsenNo ratings yet
- 22MCA1008 - Varun ML LAB ASSIGNMENTSDocument41 pages22MCA1008 - Varun ML LAB ASSIGNMENTSS Varun (RA1931241020133)100% (1)
- Experiment-7: Implementation of K-Means Clustering AlgorithmDocument3 pagesExperiment-7: Implementation of K-Means Clustering Algorithm19-361 Sai PrathikNo ratings yet
- ResNet50 Training CodeDocument9 pagesResNet50 Training Codenatashamarie.relampagosNo ratings yet
- Spark BeleskeDocument21 pagesSpark BeleskeSlavimirVesićNo ratings yet
- CRT2 LDA AssignmentDocument4 pagesCRT2 LDA AssignmentrasaramanNo ratings yet
- Midterm 2 CodesDocument15 pagesMidterm 2 CodessameertardaNo ratings yet
- Sentiment Analysis On TweetsDocument2 pagesSentiment Analysis On TweetsvikibytesNo ratings yet
- Maxbox Starter96 CNN EvaluationDocument7 pagesMaxbox Starter96 CNN EvaluationMax KleinerNo ratings yet
- Lab - Batch Data Ingestion With DMS - Instructor SetupDocument16 pagesLab - Batch Data Ingestion With DMS - Instructor SetupJob Llanos MontaldoNo ratings yet
- 2020BIT007 Assignment No7.Ipynb - ColaboratoryDocument2 pages2020BIT007 Assignment No7.Ipynb - Colaboratoryaqonline906No ratings yet
- HANDONDocument6 pagesHANDONDora Cecilia Bernal PuentesNo ratings yet
- ESTIVEN - HURTADO.SANTOS - Analytics, De, Data, No, Estructurada - Machine, Learning - ESTIVEN - HURTADO.SANTOS - Ipynb - ColaboratoryDocument5 pagesESTIVEN - HURTADO.SANTOS - Analytics, De, Data, No, Estructurada - Machine, Learning - ESTIVEN - HURTADO.SANTOS - Ipynb - ColaboratoryEstiven Hurtado SantosNo ratings yet
- Yolo v4Document42 pagesYolo v4Deepak SaxenaNo ratings yet
- SKB BuffersDocument32 pagesSKB BuffersKvivek VivekNo ratings yet
- 20bcs7635-EXP 10Document5 pages20bcs7635-EXP 10sameerNo ratings yet
- NER LabDocument65 pagesNER LabKondo CandiNo ratings yet
- Nouveau Document TexteDocument14 pagesNouveau Document TexteMANAÏ BelighNo ratings yet
- Tugas Clustering - 132021012 - Kevin Gazkia NaufalDocument6 pagesTugas Clustering - 132021012 - Kevin Gazkia NaufalkevinNo ratings yet
- ML Lab ProgramsDocument23 pagesML Lab ProgramsRoopa 18-19-36No ratings yet
- Sunbird ED Developer Bootcamp 2023Document9 pagesSunbird ED Developer Bootcamp 2023Dhamodharan ThangavelNo ratings yet
- Data Analysis and Evaluation Methods ComparisonDocument11 pagesData Analysis and Evaluation Methods ComparisonJelena NađNo ratings yet
- Clonamos El Repositorio para Obtener Los Dataset: From ImportDocument23 pagesClonamos El Repositorio para Obtener Los Dataset: From ImportJuan Chavarria AsparrinNo ratings yet
- TimeDocument215 pagesTimejamesNo ratings yet
- Deep Learning With Python FileDocument22 pagesDeep Learning With Python FileArnav ShrivastavaNo ratings yet
- BuffDocument11 pagesBuffMKNo ratings yet
- K MeansDocument4 pagesK Meansmohamed mohsenNo ratings yet
- TT - Ipynb - ColaboratoryDocument3 pagesTT - Ipynb - Colaboratoryhos1999moh78No ratings yet
- RecorDocument6 pagesRecorHariharan.kNo ratings yet
- Tugas Mandiri Pertemuan 9 - Dea Ashari Oktavia - ITSDocument1 pageTugas Mandiri Pertemuan 9 - Dea Ashari Oktavia - ITSDeaNo ratings yet
- Roop Unleashed 02.ipynbDocument15 pagesRoop Unleashed 02.ipynbeternalsoldiergirlNo ratings yet
- Maxbox Starter60 Machine LearningDocument8 pagesMaxbox Starter60 Machine LearningMax KleinerNo ratings yet
- Signal Energy and PowerDocument19 pagesSignal Energy and PowerVarsha MoharanaNo ratings yet
- Synopsis of The Transient Solvers in ESATANDocument4 pagesSynopsis of The Transient Solvers in ESATANP_leeNo ratings yet
- 2017 Fall ME349 03 NumAnalysis1Document41 pages2017 Fall ME349 03 NumAnalysis1Abdo SalahNo ratings yet
- FCM - Zip Fuzzy C - Means Clustering MATLAB, Which Contains 10 Function WWW - PudnDocument2 pagesFCM - Zip Fuzzy C - Means Clustering MATLAB, Which Contains 10 Function WWW - PudnArmando ChachiLankx Evol AlletsserhcNo ratings yet
- CSC-411-AI-lec6-Adversarial SearchDocument38 pagesCSC-411-AI-lec6-Adversarial SearchAbdullahNo ratings yet
- C++ Classes and Data Structures: Hash TablesDocument238 pagesC++ Classes and Data Structures: Hash TablesBala GanabathyNo ratings yet
- Topic4 2Document65 pagesTopic4 2YoftahiNo ratings yet
- Comm II Tutorial Sheet 1Document10 pagesComm II Tutorial Sheet 1Toka AliNo ratings yet
- Echo Cancellation in Audio Signal Using LMS Algorithm: Sanjay K. Nagendra Vinay Kumar.S.BDocument5 pagesEcho Cancellation in Audio Signal Using LMS Algorithm: Sanjay K. Nagendra Vinay Kumar.S.BPrabira Kumar SethyNo ratings yet
- Unit - 3 RS PDFDocument16 pagesUnit - 3 RS PDFSanthosh GivariNo ratings yet
- Numerical Simulation For The Solution of Nonlinear Jaulent-Miodek Coupled Equations Using Quartic B-SplineDocument18 pagesNumerical Simulation For The Solution of Nonlinear Jaulent-Miodek Coupled Equations Using Quartic B-SplineWaleed AdelNo ratings yet
- Experiment No. 3 - Roots of Equations Bracket MethodsDocument28 pagesExperiment No. 3 - Roots of Equations Bracket MethodsCedric Dela CruzNo ratings yet
- Job Sequencing With The DeadlineDocument5 pagesJob Sequencing With The DeadlineAbhiNo ratings yet
- Radix SortDocument5 pagesRadix SortMohd armanNo ratings yet
- VLSI Design of Half-Band IIR Interpolation and Decimation FilterDocument7 pagesVLSI Design of Half-Band IIR Interpolation and Decimation FiltertansnvarmaNo ratings yet
- CRC and Hamming CodeDocument12 pagesCRC and Hamming CodeAyush Raj100% (1)
- 9-Int Array Searching - SortingDocument11 pages9-Int Array Searching - SortingMuhammad Awais GhafoorNo ratings yet
- BDocument2 pagesBAhmed MalikNo ratings yet
- Beyon BroydenDocument25 pagesBeyon BroydenChristian VerdeNo ratings yet
- 110sort Malik Ch10Document81 pages110sort Malik Ch10sachinsr099No ratings yet
- Q. No. Description Question Choices Unit I-Introduction: 1 1 EasyDocument15 pagesQ. No. Description Question Choices Unit I-Introduction: 1 1 EasyArchana SainiNo ratings yet
- IJE - Volume 32 - Issue 10 - Pages 1464-1479Document16 pagesIJE - Volume 32 - Issue 10 - Pages 1464-1479IMANE TORBINo ratings yet
- 03 Unit Three orDocument61 pages03 Unit Three orAbdi Mucee TubeNo ratings yet
- Regularized Target Encoding Outperforms Traditional Methods in Supervised Machine Learning With High Cardinality FeaturesDocument22 pagesRegularized Target Encoding Outperforms Traditional Methods in Supervised Machine Learning With High Cardinality FeaturesmarboeNo ratings yet
- EC6405-Control Systems EngineeringDocument17 pagesEC6405-Control Systems EngineeringAjju K AjjuNo ratings yet
- CS1010 ETutorial 6 SolutionDocument3 pagesCS1010 ETutorial 6 SolutionYu Shu HearnNo ratings yet
- Full Name: Lab Section: ECE 3500 (Spring 2017) - Examples #1Document11 pagesFull Name: Lab Section: ECE 3500 (Spring 2017) - Examples #1Stacey BoylanNo ratings yet
- Solution of Nonlinear Equations: 1 BisectionDocument9 pagesSolution of Nonlinear Equations: 1 BisectionJose D CostaNo ratings yet
- Cisco Support Community - Upgrading Ws-3560g-24ps Ios To c3560-Ipbasek9-Mz.122-55.Se10.Bin - 2015-02-24Document2 pagesCisco Support Community - Upgrading Ws-3560g-24ps Ios To c3560-Ipbasek9-Mz.122-55.Se10.Bin - 2015-02-24Leyenda HeroeNo ratings yet
- BCS-042 Solved Assignment 042 Solved Assignment 042 Solved AssignmentDocument28 pagesBCS-042 Solved Assignment 042 Solved Assignment 042 Solved AssignmentNaksh Mayank100% (1)