Professional Documents
Culture Documents
Breast Cancer Classification Using DTC
Uploaded by
tarunbinjadagi18Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Breast Cancer Classification Using DTC
Uploaded by
tarunbinjadagi18Copyright:
Available Formats
In [1]: import pandas as pd
import warnings
warnings.filterwarnings('ignore')
from sklearn.datasets import load_breast_cancer
cancer_data = load_breast_cancer()
In [2]: list(cancer_data.target_names)
Out[2]: ['malignant', 'benign']
In [3]: cancer_data.feature_names
Out[3]: array(['mean radius', 'mean texture', 'mean perimeter', 'mean area',
'mean smoothness', 'mean compactness', 'mean concavity',
'mean concave points', 'mean symmetry', 'mean fractal dimension',
'radius error', 'texture error', 'perimeter error', 'area error',
'smoothness error', 'compactness error', 'concavity error',
'concave points error', 'symmetry error',
'fractal dimension error', 'worst radius', 'worst texture',
'worst perimeter', 'worst area', 'worst smoothness',
'worst compactness', 'worst concavity', 'worst concave points',
'worst symmetry', 'worst fractal dimension'], dtype='<U23')
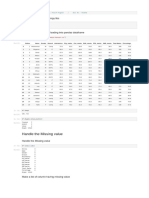
In [4]: cancer_df = pd.DataFrame(cancer_data.data, columns=cancer_data.feature_names)
cancer_df.head()
Out[4]:
mean mean worst worst
mean mean mean mean mean mean mean mean worst worst worst worst worst worst worst worst
concave fractal ... concave fractal
radius texture perimeter area smoothness compactness concavity symmetry radius texture perimeter area smoothness compactness concavity symmetry
points dimension points dimension
0 17.99 10.38 122.80 1001.0 0.11840 0.27760 0.3001 0.14710 0.2419 0.07871 ... 25.38 17.33 184.60 2019.0 0.1622 0.6656 0.7119 0.2654 0.4601 0.11890
1 20.57 17.77 132.90 1326.0 0.08474 0.07864 0.0869 0.07017 0.1812 0.05667 ... 24.99 23.41 158.80 1956.0 0.1238 0.1866 0.2416 0.1860 0.2750 0.08902
2 19.69 21.25 130.00 1203.0 0.10960 0.15990 0.1974 0.12790 0.2069 0.05999 ... 23.57 25.53 152.50 1709.0 0.1444 0.4245 0.4504 0.2430 0.3613 0.08758
3 11.42 20.38 77.58 386.1 0.14250 0.28390 0.2414 0.10520 0.2597 0.09744 ... 14.91 26.50 98.87 567.7 0.2098 0.8663 0.6869 0.2575 0.6638 0.17300
4 20.29 14.34 135.10 1297.0 0.10030 0.13280 0.1980 0.10430 0.1809 0.05883 ... 22.54 16.67 152.20 1575.0 0.1374 0.2050 0.4000 0.1625 0.2364 0.07678
5 rows × 30 columns
In [5]: from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(cancer_df,
cancer_data.target,
test_size = 0.33,
random_state = 42)
In [6]: from sklearn.tree import DecisionTreeClassifier
from sklearn import metrics
In [7]: # Create Decision Tree classifer object
dtc = DecisionTreeClassifier()
# Train Decision Tree Classifer
model = dtc.fit(X_train,y_train)
#Predict the response for test dataset
y_pred = model.predict(X_test)
In [8]: # Model Accuracy, how often is the classifier correct?
print("Accuracy:",metrics.accuracy_score(y_test, y_pred))
Accuracy: 0.925531914893617
In [9]: from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, y_pred)
Out[9]: array([[ 63, 4],
[ 10, 111]], dtype=int64)
In [10]: import matplotlib.pyplot as plt
from sklearn.metrics import plot_confusion_matrix
matrix = plot_confusion_matrix(model, X_test, y_test, cmap=plt.cm.Reds)
In [11]: from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
precision recall f1-score support
0 0.85 0.91 0.88 67
1 0.95 0.91 0.93 121
accuracy 0.91 188
macro avg 0.90 0.91 0.90 188
weighted avg 0.91 0.91 0.91 188
You might also like
- ML 4Document4 pagesML 4yefigoh133No ratings yet
- Cancer de Mama Sin Estandarizar EstratificadoDocument10 pagesCancer de Mama Sin Estandarizar EstratificadoJIMMY CRISTHIAN CJURO APAZANo ratings yet
- 20BCP021 Assignment 3Document7 pages20BCP021 Assignment 3chatgptplus4usNo ratings yet
- From Import Import As From Import From Import From Import From Import From Import Import From Import ImportDocument5 pagesFrom Import Import As From Import From Import From Import From Import From Import Import From Import ImportNguyen Trung ThinhNo ratings yet
- 13 Cross - ValidationDocument4 pages13 Cross - ValidationGabriel GheorgheNo ratings yet
- Breast Cancer DatasetDocument154 pagesBreast Cancer DatasetIris RumiNo ratings yet
- 1Document4 pages1Arpita DasNo ratings yet
- Fashion MNIST-6Document10 pagesFashion MNIST-6akif barbaros dikmenNo ratings yet
- 1FsWES7YJDERHD-bZ2ujFakbQyzi6 YinDocument9 pages1FsWES7YJDERHD-bZ2ujFakbQyzi6 YinNoel BinoyNo ratings yet
- Support Vector Machine (SVM) - BioinformaticsDocument10 pagesSupport Vector Machine (SVM) - BioinformaticsalihamzakrbgNo ratings yet
- K Nearest NeighboursDocument4 pagesK Nearest Neighboursbunsglazing135No ratings yet
- ANANYAA GUPTA 20BCT0177 ML MTT 24/11/21 Q3 Breast Cancer DatasetDocument4 pagesANANYAA GUPTA 20BCT0177 ML MTT 24/11/21 Q3 Breast Cancer DatasetAnanyaa GuptaNo ratings yet
- 2 and 3Document6 pages2 and 3Radhika KhandelwalNo ratings yet
- Random Forest: Implementaciones de Scikit-Learn Sobre QSARDocument11 pagesRandom Forest: Implementaciones de Scikit-Learn Sobre QSARRichard Jimenez100% (1)
- Image ClassificationsDocument4 pagesImage ClassificationsAnvesh SilaganaNo ratings yet
- Project SVMDocument16 pagesProject SVMCalo SoftNo ratings yet
- Practical - 5 - 52Document4 pagesPractical - 5 - 52Royal EmpireNo ratings yet
- Assignment 11Document7 pagesAssignment 11ankit mahto100% (1)
- Meaningful Predictive Modeling Week-4 Assignment Cancer Disease PredictionDocument6 pagesMeaningful Predictive Modeling Week-4 Assignment Cancer Disease PredictionfrankhNo ratings yet
- 4.4. Data Standardization - Ipynb - ColaboratoryDocument1 page4.4. Data Standardization - Ipynb - Colaboratorylokesh kNo ratings yet
- Assignment-7 (CSL5402) Name:-Kommireddy - Manikanta Sai Roll No: - 1906036 Branch: - CSE-1Document7 pagesAssignment-7 (CSL5402) Name:-Kommireddy - Manikanta Sai Roll No: - 1906036 Branch: - CSE-1Manikanta Sai100% (1)
- Assignment-7 (CSL5402) Name:-Kommireddy - Manikanta Sai Roll No: - 1906036 Branch: - CSE-1Document7 pagesAssignment-7 (CSL5402) Name:-Kommireddy - Manikanta Sai Roll No: - 1906036 Branch: - CSE-1Manikanta Sai100% (1)
- BreastcancerDocument13 pagesBreastcancergntfhx9pydNo ratings yet
- Task 1Document9 pagesTask 1Dương Vũ MinhNo ratings yet
- RAJIV RANJAN - 19!02!2023 - Predictive Modelling Project Report - FinalDocument34 pagesRAJIV RANJAN - 19!02!2023 - Predictive Modelling Project Report - FinalRajiv Ranjan100% (1)
- Template For CH 6-9Document22 pagesTemplate For CH 6-9prabhjot kaurNo ratings yet
- Loading The Dataset: 'Diabetes - CSV'Document4 pagesLoading The Dataset: 'Diabetes - CSV'Divyani ChavanNo ratings yet
- Recurring Neural Networks For Sequence - Sentiment Analysis With The IMDb Dataset - Ipynb - ColaboratoryDocument16 pagesRecurring Neural Networks For Sequence - Sentiment Analysis With The IMDb Dataset - Ipynb - Colaboratoryfardowsa mohamedNo ratings yet
- Breast Cancer Classification With Machine LearningDocument17 pagesBreast Cancer Classification With Machine LearningAiza EmaanNo ratings yet
- Loading The Dataset: 'Churn - Modelling - CSV'Document6 pagesLoading The Dataset: 'Churn - Modelling - CSV'Divyani ChavanNo ratings yet
- Dsbda 4Document4 pagesDsbda 4Arbaz ShaikhNo ratings yet
- 3 Confussion Matrix Hasil Modelling OKDocument8 pages3 Confussion Matrix Hasil Modelling OKArman Maulana Muhtar100% (1)
- Elementary Statistics 6th Edition Larson Solutions Manual 1Document47 pagesElementary Statistics 6th Edition Larson Solutions Manual 1suzanne100% (32)
- Elementary Statistics 6Th Edition Larson Solutions Manual Full Chapter PDFDocument36 pagesElementary Statistics 6Th Edition Larson Solutions Manual Full Chapter PDFshelia.hamada995100% (12)
- 7.01 Feature SelectionDocument3 pages7.01 Feature Selectionjayant khanvilkarNo ratings yet
- Covid-19 Prediction - Jupyter NotebookDocument6 pagesCovid-19 Prediction - Jupyter NotebookTEEGALAMEHERRITHWIK 122010320035No ratings yet
- NguyenTrungThinh BT3.3Document5 pagesNguyenTrungThinh BT3.3Nguyen Trung ThinhNo ratings yet
- Compressing Polygon Mesh Geometry: With Parallelogram PredictionDocument80 pagesCompressing Polygon Mesh Geometry: With Parallelogram PredictionAntonio Carlos Salzvedel FurtadoNo ratings yet
- Machine Learnin1Document41 pagesMachine Learnin1Surabhi Kulkarni100% (1)
- Chapter 9: Selection of VariablesDocument30 pagesChapter 9: Selection of VariablesMerk GetNo ratings yet
- Khoirul Presentasi GADocument47 pagesKhoirul Presentasi GAIwan JuandaNo ratings yet
- Elementary Statistics 6th Edition Larson Solutions Manual 1Document36 pagesElementary Statistics 6th Edition Larson Solutions Manual 1cynthiafrankgetbzdpkyf100% (22)
- Project Coding-Manish Dwari 1807Document1 pageProject Coding-Manish Dwari 1807soniakar3000No ratings yet
- Mloa Exp1 C121Document49 pagesMloa Exp1 C121Devanshu MaheshwariNo ratings yet
- Testing If A Relationship Occurs Between Two Variables Using CorrelationDocument26 pagesTesting If A Relationship Occurs Between Two Variables Using CorrelationKrishna KantNo ratings yet
- Lab - 5 (CB - En.u4ece22115)Document5 pagesLab - 5 (CB - En.u4ece22115)Daejuswaram GopinathNo ratings yet
- LightGBM - An In-Depth Guide PythonDocument26 pagesLightGBM - An In-Depth Guide PythonRadha KilariNo ratings yet
- TorsionDocument6 pagesTorsionnasirhakimiNo ratings yet
- Lab10 Regression Evaluation MethodsDocument5 pagesLab10 Regression Evaluation Methodsiffi khanNo ratings yet
- Practical-5 - Jupyter NotebookDocument8 pagesPractical-5 - Jupyter NotebookHarsha Gohil100% (1)
- ML0101EN Clas Logistic Reg Churn Py v1Document13 pagesML0101EN Clas Logistic Reg Churn Py v1banicx100% (1)
- Bending Moment&SheerForce ExperimentDocument14 pagesBending Moment&SheerForce ExperimentZhenyi OoiNo ratings yet
- CatBoost - An In-Depth Guide PythonDocument33 pagesCatBoost - An In-Depth Guide PythonRadha KilariNo ratings yet
- SVM (Support Vector Machine) For Classification - by Aditya Kumar - Towards Data ScienceDocument28 pagesSVM (Support Vector Machine) For Classification - by Aditya Kumar - Towards Data ScienceZuzana W100% (1)
- Fault Detection Using Data Based ModelsDocument14 pagesFault Detection Using Data Based ModelsPierpaolo VergatiNo ratings yet
- Iv Edit 2Document29 pagesIv Edit 2ardat ahmadNo ratings yet
- Assignment 1 - Introduction To Machine Learning: Version 1.0 of This Notebook. To DownloadDocument30 pagesAssignment 1 - Introduction To Machine Learning: Version 1.0 of This Notebook. To DownloadJayanth0% (1)
- Handle The Missing Value: Importing Pandas and Numpy LibsDocument4 pagesHandle The Missing Value: Importing Pandas and Numpy LibsAkshay PatilNo ratings yet
- 3d OpticsDocument16 pages3d OpticsKranthi LakumNo ratings yet
- Indian Perspective For Probiotics: A Review: ArticleDocument12 pagesIndian Perspective For Probiotics: A Review: ArticleMurali DathanNo ratings yet
- Health Pass SertifikaDocument2 pagesHealth Pass SertifikaJeyhun RamazanovNo ratings yet
- Uhms DCS Age GuidelinesDocument17 pagesUhms DCS Age GuidelinesMarios PolyzoisNo ratings yet
- JaundiceDocument19 pagesJaundiceketsela12No ratings yet
- Funda Lab - Prelim ReviewerDocument16 pagesFunda Lab - Prelim ReviewerNikoruNo ratings yet
- Oxygen TherapyDocument31 pagesOxygen TherapyChayan BhowmikNo ratings yet
- Module 4: Mood Disorders: Disorders and Individuals With Bipolar Disorders. The Key Difference BetweenDocument7 pagesModule 4: Mood Disorders: Disorders and Individuals With Bipolar Disorders. The Key Difference BetweenLiz Evermore0% (1)
- Collection of Past Papers by Angina Pectoris PDFDocument79 pagesCollection of Past Papers by Angina Pectoris PDFMuhammad Omar Akram100% (2)
- Crowns and Other Extra-Coronal RestorationDocument116 pagesCrowns and Other Extra-Coronal RestorationDennis La Torre Zea100% (4)
- NCP For RDSDocument3 pagesNCP For RDSKevin P. Feliciano74% (23)
- Heavy Menstrual Bleeding Assessment and Management PDF 1837701412549Document33 pagesHeavy Menstrual Bleeding Assessment and Management PDF 1837701412549Meera Al AliNo ratings yet
- Literature Review 1Document5 pagesLiterature Review 1api-550490262No ratings yet
- Immunization Schedule in India 2017 (Latest !!)Document13 pagesImmunization Schedule in India 2017 (Latest !!)rajNo ratings yet
- Teleradiologi: Cross Reporting & Smart AssignDocument17 pagesTeleradiologi: Cross Reporting & Smart Assignrafael100% (1)
- Chap1 Eks 2ed AddendumDocument19 pagesChap1 Eks 2ed AddendumCamroc BraineryNo ratings yet
- McCracken Diseno Actual de PPRDocument9 pagesMcCracken Diseno Actual de PPRRania LVNo ratings yet
- Epker y Fish. 1978. Corrección QX de ADF PDFDocument17 pagesEpker y Fish. 1978. Corrección QX de ADF PDFERICK FABIAN FAJARDO NORATONo ratings yet
- CH 29Document39 pagesCH 29Jann Zaniel Allayne RiNo ratings yet
- Vaginal MicrobiomeDocument9 pagesVaginal MicrobiomeRika Yulizah GobelNo ratings yet
- Avenir Hip System Surgical TechniqueDocument16 pagesAvenir Hip System Surgical Techniquesuraj mahadikNo ratings yet
- Clinical Evaluation Report For Trilogy Evo Ventilator: Prepared For: Respironics, IncDocument79 pagesClinical Evaluation Report For Trilogy Evo Ventilator: Prepared For: Respironics, Incdaphnestc0211No ratings yet
- SA Anesthesia Drug Dosages 2022Document3 pagesSA Anesthesia Drug Dosages 2022KRLITHIU Borja100% (1)
- CM Sputum Examination Ocfemia Eliazel G.Document41 pagesCM Sputum Examination Ocfemia Eliazel G.eliazel ocfemiaNo ratings yet
- Prudential BSN Takaful Berhad Investment-Linked Plan IllustrationDocument19 pagesPrudential BSN Takaful Berhad Investment-Linked Plan IllustrationhilmiyaidinNo ratings yet
- Geriatric Physical TherapyDocument6 pagesGeriatric Physical Therapyirdasriwahyuni0% (1)
- Re EvaluationDocument4 pagesRe EvaluationchandanaNo ratings yet
- Marijuana: A Medicine For CancerDocument11 pagesMarijuana: A Medicine For CancerXylvia Hannah Joy CariagaNo ratings yet
- 0 - Review of Post Graduate Medical Entrance Examination (PGMEE) (AAA) (PDFDrive - Com) Export PDFDocument39 pages0 - Review of Post Graduate Medical Entrance Examination (PGMEE) (AAA) (PDFDrive - Com) Export PDFAbu ÂwŞmž100% (2)
- Pathophysiology QBDocument5 pagesPathophysiology QBVimlesh PalNo ratings yet
- Pre Dent Ate Period PedoDocument16 pagesPre Dent Ate Period PedoFourthMolar.com0% (1)