0% found this document useful (0 votes)
36 views14 pagesUnderstanding miRNA and siRNA Functions
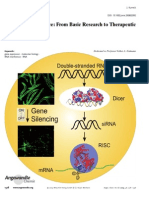
The document discusses the roles of miRNA and siRNA in RNA interference, a natural process that silences gene expression by degrading mRNA. It highlights the mechanisms of action for both siRNA and miRNA, including their processing, target recognition, and therapeutic potential. Additionally, it addresses the impact of viral suppressors on RNA silencing and provides resources for siRNA design and validation tools.
Uploaded by
iemanasimjkpCopyright
© © All Rights Reserved
We take content rights seriously. If you suspect this is your content, claim it here.
Available Formats
Download as PPTX, PDF, TXT or read online on Scribd
0% found this document useful (0 votes)
36 views14 pagesUnderstanding miRNA and siRNA Functions
The document discusses the roles of miRNA and siRNA in RNA interference, a natural process that silences gene expression by degrading mRNA. It highlights the mechanisms of action for both siRNA and miRNA, including their processing, target recognition, and therapeutic potential. Additionally, it addresses the impact of viral suppressors on RNA silencing and provides resources for siRNA design and validation tools.
Uploaded by
iemanasimjkpCopyright
© © All Rights Reserved
We take content rights seriously. If you suspect this is your content, claim it here.
Available Formats
Download as PPTX, PDF, TXT or read online on Scribd