Professional Documents
Culture Documents
Microbiology Bacteria Identification
Uploaded by
nabilahOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Microbiology Bacteria Identification
Uploaded by
nabilahCopyright:
Available Formats
EXPERIMENT 4: BACTERIA IDENTIFICATION
UNIVERSITY KUALA LUMPUR MALAYSIAN INSTITUTE OF
CHEMICAL AND BIOENGINEERING TECHNOLOGY (MICET)
LAB REPORT MICROBIAL TECHNOLOGY
EXPERIMENT 4: BACTERIAL IDENTIFICATION
PREPARED BY: DAYANG NURSYAZA FAZLIN BINTI AWANG KEDERI
ID: 55211114328
CHECKED BY:
DR LEONG CHEAN RING
DATE OF SUBMISSION:
MARCH 18, 2016
EXPERIMENT 4: BACTERIA IDENTIFICATION
OBJECTIVES
To identify bacteria by conducting series of biochemical test
To practise 5Is which are Inoculation, Incubation, Isolation, Inspection and
Identification
INTRODUCTION
Biochemical tests are the tests used for the identification of bacteria species based on
the differences in the biochemical activities of different bacteria. .Bacterial physiology differs
from one species to the other. These differences in carbohydrate metabolism, protein
metabolism, fat metabolism, production of certain enzymes, ability to utilize a particular
compound help them to be identified by the biochemical tests.
The identification of higher plants and animals involves the observations of the
structural differences, both internal and external, which exist among them. Most of these
structural differences are visible to naked eye. Even the identification of microscopic plants
and animals involves the observations of their structural differences under a microscope.Such
identification based on structural differences is not possible in case of bacteria, because
structural differences, which may differentiate one species of bacteria from the other, are not
discernible even under a microscope. The structural differences with respect to shape, size
and arrangement of bacteria only help in the process of identification, because there are many
species of bacteria having similar shape, size and arrangement.
Therefore, ultimately, the identification of bacteria is mostly based on the differences
in their biochemical activities. There are so many examples of biochemical tests but in this
experiment, the tests that been used are only few which are Starch Hydrolysis test, Methyl
Red test, Voges-Proskauer test, Citrate utilisation test and differential on MacConkey Agar.
Citrate utilisation test is defined as a medium used to determine if an organism can
use citrate as its sole carbon source. It is often used to differentiate between members
ofEnterobacteriaceae. In organisms capable of utilizing citrate as a carbon source, the
enzyme citrase hydrolyzes citrate into oxaoloacetic acid and acetic acid. The oxaloacetic acid
is then hydrolyzed into pyruvic acid and CO 2. If CO2 is produced, it reacts with components
EXPERIMENT 4: BACTERIA IDENTIFICATION
of the medium to produce an alkaline compound (e.g. Na 2CO3). The alkaline pH turns the pH
indicator (bromthymol blue) from green to blue.
Methyl red test is used to determine which fermentation pathway is used to utilize
glucose. In the mixed acid fermentation pathway, glucose is fermented and produces several
organic acids (lactic, acetic, succinic, and formic acids). The stable production of enough acid
to overcome the phosphate buffer will result in a pH of below 4.4. If the pH indicator (methyl
red) is added to an aliquot of the culture broth and the pH is below 4.4, a red color will
appear. If the MR turns yellow, the pH is above 6.0 and the mixed acid fermentation pathway
has not been utilized The Voges-Proskauer test detects the presence of acetoin, a precursor of
2, 3 butanediol. If the culture is positive for acetoin, it will turn brownish-red to pink. If the
culture is negative for acetoin, it will turn brownish-green to yellow (Note: A culture will
usually only be positive for one pathway: either MR+ or VP+. Escherichia coli is MR+ and
VP-)
Meanwhile MacConkey Agar is a medium which is both selective and differential.
The selective ingredients are the bile salts and the dye, crystal violet which inhibit the growth
of Gram-positive bacteria. The differential ingredient is lactose. Fermentation of this sugar
results in an acidic pH and causes the pH indicator, neutral red, to turn a bright pinky-red
color thus, the organisms that capable in lactose fermentation such as Escherichia coli, form
bright pinky-red colonies. MacConkey agar is commonly used to differentiate between the
Enterobacteriaceae.
Lastly, the starch hydrolysis test is where the known starch is immunized with the
given unknown species. After being immunized, Iodine is added to the mixtures and which
shows if any starch is present in the mixture or not. If Iodine turns blue-black then that means
there is starch present in the mixture and if not then there is no starch.
Last but not least, aseptic technique is practised in every test to prevent contamination
of other unwanted organism.
EXPERIMENT 4: BACTERIA IDENTIFICATION
SUMMARY
This experiment was conducted to identify bacteria by using biochemical test. There
are five (5) biochemical tests which are Starch Hydrolysis test, Methyl Red test, VogesProskauer test, Citrate utilisation test and differential on MacConkey Agar. For Starch
Hydrolysis test and MacConkey Agar test, B. subtillis and E.Coli were inoculated and place
on the Starch agar. Before that, the agar has been divided by two section by using marker pen.
They are then incubated for 24 hours at 37 . For the Starch agar, after incubated, iodine was
poured evenly then observed. While for the MacConkey Agar, the plate was observed directly
without any further procedure. For the Methyl Red and Vogue-Proskauer, the colony (B.
subtillis and E.Coli) were transferred respectively into the glucose phosphate broth and
incubate at 37 C for 24 h. After that for Methyl Red, 2 drops of methyl red solution into the
culture is added after 24 hours of incubation and the colour change is observed. Meanwhile
for the Vogue-Proskauer, 2 drops (about 0.5 ml) of creatine solution and 1 ml of 4%
potassium hydroxide solution is added and the colour change is observed after 4 hours. An
aseptic technique is practised in every test to avoid contamination of the other organisms by
using flame sterilization and 70% of ethanol.
EXPERIMENT 4: BACTERIA IDENTIFICATION
EXPERIMENTAL PROCEDURE
(A) Growth on the selective and differential MacConkey agar
1. One plate of MacConkey agar is taken and a line is drawed by using marker pen to
separate it into two zones: A and B.
2. Both B. subtillis and E. coli is inoculated on zone A and B respectively
3. The plates are incubated at 37 C for 24 hours.
4. The growth of both bacteria is observed on the plate.
(B) Starch Hydrolysis Test
1. One plate of starch agar is taken and a line is drawed using marker pen to separate it into
two zones: A and B.
2. Both B. subtillis and E. coli is inoculated on zone A and B respectively
3. The plates are incubated at 37 C for 24 hours.
4. Iodine is poured over the surface of the starch plate to cover the entire plate. The plate is
rotated and tipped to spread the iodine.
5. The bacteria shows positive result is observed. (Shown by production of a clear zone
around the bacteria growth.
(C) Methyl red (2 test tubes containing 5 ml of phosphate broth + 0.5% (w/v)
glucose)
1. Using the inoculating wire loop, the colony is transferred into the glucose phosphate broth
and incubated at 37 C for 24 h.
2. After 24 h, 2 drops of methyl red solution is added into the culture. The mixture is shakes
gently and the final colour change is observed. Red indicates positive result.
EXPERIMENT 4: BACTERIA IDENTIFICATION
(D) Voges Proskauer (2 test tubes containing 5 ml of phosphate broth + 0.5% (w/v)
glucose)
1. Using the inoculating wire loop, the colony is transferred into the glucose phosphate
broth and incubated at 37 C for 24 h.
2. After 24 h, 2 drops (about 0.5 ml) of creatine solution is added and 1 ml of 4%
potassium hydroxide solution is pippeted. The mixture is shaken and after 1-4 h the
result is examined. A positive reaction is indicated by a change of colour from yellow
to pink.
(E) Citrate utilization test (2 Universal bottles containing Simmon- citrate agar
slants)
1. Using the inoculating wire loop, each of the colonies is streaked onto the slant of
Simmon-citrate medium and incubated at 37 C for 24 h.
2. After 24 h, the colour change is examined and growth of the microorganisms is observed.
Utilization of citrate is indicated by the colour changes from green to blue.
EXPERIMENT 4: BACTERIA IDENTIFICATION
DATA AND RESULTS
(A) Growth on the selective and differential MacConkey agar
Figure 1: MacConkey Agar test for B. subtillis (A zone) and E. Coli (B zone)
Observation: negative MacConkey Agar test for and positive MacConkey Agar test for E. Coli
There is growth of E. Coli on zone B and around of it has a pink clear zone
There is no growth of B. Subtillis on zone A whereas the zone is clear.
(B)Starch Hydrolysis Test
EXPERIMENT 4: BACTERIA IDENTIFICATION
Figure 2: Starch Hydrolysis test for B. subtillis (A zone) and E. Coli (B zone)
Observation: - zone of clearing for B. Subtillis is positive while for E. Coli is negative
(C) Methyl red (2 test tubes containing 5 ml of phosphate broth + 0.5% (w/v) glucose)
Figure 3: Methyl Red test for B. subtillis (right) and E. Coli (left)
Observation:1. The right bottle which is E. Coli turn reddish
2. The left bottle which is B. Subtillis has no change
(D) Voges Proskauer (2 test tubes containing 5 ml of phosphate broth + 0.5% (w/v)
glucose
EXPERIMENT 4: BACTERIA IDENTIFICATION
Figure 4: Voges Proskauer test for B. subtillis (right) and E. Coli (left)
Observation:- Both are negative results for the color change.
(E) Citrate utilization test (2 Universal bottles containing Simmon- citrate agar
slants)
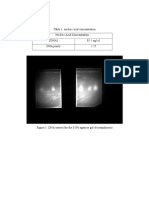
Figure 5: Citrate utilization test for E. Coli (left) and B. subtillis (right)
Observation: - E. Coli shows Citrate negative test and B. subtillis is positive Citrate test,
which turns green to blue.
DISCUSSION
9
EXPERIMENT 4: BACTERIA IDENTIFICATION
The microbiology lab involved the inoculating many differential media including
MacConkey Agar, Starch hydrolysis agar, Voges Proskauer broth, Methyl Red broth and
Simmons Citrate Agar slant. Each media had a distinct purpose, some even tested for more
than one characteristic. Results were observed after 24 hours of incubation and are interpreted
in this paragraph along with the explanations of the workings of each media type used in the
lab.
First to be discussed is the MacConkey Agar result. MacConkey agar was determined
to be selective because it prevented the growth of gram positive bacteria by means of bile
salts and crystal violet dye. The pink coloration of the agar came from the crystal violet.
MacConkey agar also included lactose, which was converted to lactid acid by lactose
fermentation. In this lab, B. Subtillis (A) shows clear zone, opaque and colorless which
indicates it is negative, thus it is not labelled as lactose fermenter while E. Coli (B) resulted in
bright-red colonies which means it is able to ferment lactose. MacConkey Agar were said to
be differential because its ability to detect the enteric pathogens and coliforms.
Next is Starch Hydrolysis tests result. Based on the results obtained, there is clear
zone at B. Subtillis (A) and no growth at all in E. Coli (B). This is because, the hydrolysis of
the starch will create a clear zone around the bacterial growth, which explain the results that
obtained at the zone of B. Subtillis (A). On top of that, in order to interpret the results of the
starch hydrolysis test, iodine must be added to the agar. It will reacts with the starch to form
dark brown color.
For the Methyl red test, is is named because it developed a red hue if acidie end
products of glucose were present. Methyl red turns red (positive result) for the E. Coli at a pH
of 4, and yellow (negative result) for the B. Subtillis at a pH of 6. These outcomes shows that
E. Coli created acidic end products in their catabolism of glucose, while B. Subtillis either did
not ferment glucose or did not produce the same amounts of acidic products.
In the Voges-Proskauer test, this test is used to determine which fermentation pathway
is used to utilize glucose. This test detects the presence of the acetoin, a precursor of 2,3
butanediol. If the culture is positive for acetoin, it will turn brownish red to pink. If the
culture is negative, it will turn brownish green to yellow. But the result that obtained in this
10
EXPERIMENT 4: BACTERIA IDENTIFICATION
experiment were both yellow in colour, which is against the Voges Proskaeur principle. This
might be because some human error that might be discussed later.
Lastly for the Simmons Citrate Agar tests, it is defined to determined bacterial
species ability to use sodium citrate as its only carbon source to produce energy. If the
species was equipped with a citrate permease to transport citrate into its cells, then the
bacteria could convert the citrate to carbon dioxide and pyruvic acid. This carbon dioxide is
then combined with Na+ from the sodium citrate to produce alkaline sodium carbonate.
Sodium carbonate production raises the pH and turns bromothymol blue from green to blue,
indicating alkalinity. Since citrate utilization requires oxygen, SCA tests are made in slant
form. Citrate negative test was shown at the E. Coli while Citrate positive test was shown at
the B. Subtillis.
Last but not least, there are few error might occurred during experiment, which
affecting the results obtained. Firstly, the streaking techniques. Although aseptic techniques
are practised along the way while conducting the experiment, it might be the steel rod, which
is used to transfer colony was still too hot, hence destroyed or killed the desired colony. Next
is, the procedure might be so simple, but since the experiment was conducted at the outside of
the fume hood, unwanted organism might contaminate the sample.
11
EXPERIMENT 4: BACTERIA IDENTIFICATION
CONCLUSION AND RECOMMENDATIONS
It can be concluded that tests used to identify Gram Positive Bacteria is Starch
Hydrolysis test while the tests that can be used to identify Gram Negative Bacteria are
Methyl Red / Voges Proskauer, MacConkey Agar and Simmons Citrate Agar. This
experiment can be said is a success even though there was some error that might be
unavoidable. The objective of the experiment which are to identify bacteria by conducting
series of biochemical test and to practise 5Is which are Inoculation, Incubation, Isolation,
Inspection and Identification is accomplished. As we observed previously, bacterial
colonies can digger greatly in their morphologies. These differences can help us
identifying different species of bacteria. Likewise, bacterial species differ in their cellular
morphologies and staining properties. Selective and differential media could be use in
identifying bacterial species for example, MacConkey Agar. Generally, selective and
differential media rely on some structural or metabolic property of the species that is
preferentially selected. The recommendation for this experiment could be, all test should
be performed in fume hood to decrease the risk of contamination and the steel rod use to
inoculate colony should be cooled down at least for few seconds, to avoid killing the
desired bacteria.
12
EXPERIMENT 4: BACTERIA IDENTIFICATION
TUTORIAL
1. You have isolated an amylase-producing bacterium from the soil sample collected
from oil palm plantation in Alor Gajah, Melaka. Propose a few alternatives to identify
the bacterial strain.
Few alternatives that can be considered are Cultural characterization where the
isolates were observed under microscope to obtain the colony morphology in example
its colour, shape, size, nature of colony and pigmentation. Next is Microscopic
observation. The bacterial isolates were gram stained and observed under a high
power magnifying lens in light microscope. Besides, screening of potent amylase
producing bacteria by starch hydrolysis test can be performed. Bacteria isolates were
screened for amylolytic activity by starch hydrolysis test on an agar starch plates. The
microbial isolates were streaked on the starch agar plate and incubated at 37C for 48
hours. After incubation iodine solution was flooded with dropper for 30 seconds on
the starch agar plate. Presence of blue color around the growth indicates negative
result and a clear zone of hydrolysis around the growth indicates positive result. The
isolates produced clear zones of hydrolysis were considered as amylase producers and
were further investigated
REFERENCES
13
EXPERIMENT 4: BACTERIA IDENTIFICATION
1. Alfred. E. Brown. (2007). Bensons microbiological applications: Laboratory
Manual in General Microbiology. (10th edition). New York: Mc Graw Hill.
2. Johnson, T.R., & Case, C.L., (2007).Laboratory Experiments in Microbiology. (8 th
Edition). San Francisco. Pearson Education.
3. Biology
Labs.
(n.d.)
Retrieved
March
http://www.odinity.com/biochemical-activities/
4. Starch Hydrolysis Test. (n.d.). Retrieved
12,
March
2016,
12,
2016,
from
from
http://www.vumicro.com/vumie/help/vumicro/Starch_Hydrolysis_Test.html
5. Citrate Utilization Test-Principle, Media, Procedure and Result. 2015. Retrieved
March 12, 2016, from http://www.microbiologyinfo.com/citrate-utilization-testprinciple-media-procedure-and-results/
14
You might also like
- Never Split the Difference: Negotiating As If Your Life Depended On ItFrom EverandNever Split the Difference: Negotiating As If Your Life Depended On ItRating: 4.5 out of 5 stars4.5/5 (838)
- Procedure MicrobDocument14 pagesProcedure MicrobnabilahNo ratings yet
- Shoe Dog: A Memoir by the Creator of NikeFrom EverandShoe Dog: A Memoir by the Creator of NikeRating: 4.5 out of 5 stars4.5/5 (537)
- JijlijolllllllllllllllDocument1 pageJijlijolllllllllllllllnabilahNo ratings yet
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeFrom EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeRating: 4 out of 5 stars4/5 (5794)
- Results DnaDocument2 pagesResults DnanabilahNo ratings yet
- The Yellow House: A Memoir (2019 National Book Award Winner)From EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Rating: 4 out of 5 stars4/5 (98)
- Aseptic Techniques Without Aseptic Techniques Nutrient Broth Nutrient Agar Nutrient Broth Nutrient Agar Growth (+) or (-)Document1 pageAseptic Techniques Without Aseptic Techniques Nutrient Broth Nutrient Agar Nutrient Broth Nutrient Agar Growth (+) or (-)nabilahNo ratings yet
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceFrom EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceRating: 4 out of 5 stars4/5 (894)
- Experiment 2Document3 pagesExperiment 2nabilah0% (1)
- The Little Book of Hygge: Danish Secrets to Happy LivingFrom EverandThe Little Book of Hygge: Danish Secrets to Happy LivingRating: 3.5 out of 5 stars3.5/5 (399)
- CLB 11103-Biology of CellDocument6 pagesCLB 11103-Biology of CellnabilahNo ratings yet
- CLB 20502-Bioscience StatisticsDocument16 pagesCLB 20502-Bioscience StatisticsnabilahNo ratings yet
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureFrom EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureRating: 4.5 out of 5 stars4.5/5 (474)
- CLB 11103 Biology of CellsDocument6 pagesCLB 11103 Biology of CellsnabilahNo ratings yet
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryFrom EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryRating: 3.5 out of 5 stars3.5/5 (231)
- CLB 11103-Biology of CellDocument6 pagesCLB 11103-Biology of CellnabilahNo ratings yet
- Grit: The Power of Passion and PerseveranceFrom EverandGrit: The Power of Passion and PerseveranceRating: 4 out of 5 stars4/5 (587)
- Chapter 1.1 Water EdDocument42 pagesChapter 1.1 Water EdnabilahNo ratings yet
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaFrom EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaRating: 4.5 out of 5 stars4.5/5 (266)
- CLB 11103 Biology of CellsDocument6 pagesCLB 11103 Biology of CellsnabilahNo ratings yet
- Experiment 1 EdDocument10 pagesExperiment 1 EdMsfaeza HanafiNo ratings yet
- On Fire: The (Burning) Case for a Green New DealFrom EverandOn Fire: The (Burning) Case for a Green New DealRating: 4 out of 5 stars4/5 (73)
- Time and Motion Study of OPDDocument15 pagesTime and Motion Study of OPDsaurabh100% (1)
- The Emperor of All Maladies: A Biography of CancerFrom EverandThe Emperor of All Maladies: A Biography of CancerRating: 4.5 out of 5 stars4.5/5 (271)
- Museum functional areas guideDocument8 pagesMuseum functional areas guideChelle CruzNo ratings yet
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersFrom EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersRating: 4.5 out of 5 stars4.5/5 (344)
- Cases Digest on Adoption, Guardianship and Related LawsDocument2 pagesCases Digest on Adoption, Guardianship and Related LawsGillian BrionesNo ratings yet
- The Unwinding: An Inner History of the New AmericaFrom EverandThe Unwinding: An Inner History of the New AmericaRating: 4 out of 5 stars4/5 (45)
- DARACLAR 9000HP Silica Brewery Case Study 2016-WebDocument7 pagesDARACLAR 9000HP Silica Brewery Case Study 2016-WebAlia ShabbirNo ratings yet
- Team of Rivals: The Political Genius of Abraham LincolnFrom EverandTeam of Rivals: The Political Genius of Abraham LincolnRating: 4.5 out of 5 stars4.5/5 (234)
- Premium detergent market insights and Nirma case studyDocument32 pagesPremium detergent market insights and Nirma case studyBhavya ShahNo ratings yet
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyFrom EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyRating: 3.5 out of 5 stars3.5/5 (2219)
- Palm Kernel Oil Mill ProjectDocument8 pagesPalm Kernel Oil Mill Projectsjr141071100% (2)
- 01 Itp-380kv Gis - PlanDocument9 pages01 Itp-380kv Gis - PlanYahya SamaraNo ratings yet
- Product RD Session 16 - Phase Eqm Part 3Document38 pagesProduct RD Session 16 - Phase Eqm Part 3Rishabh JainNo ratings yet
- MONO POWER AMPLIFIER SERVICE MANUALDocument34 pagesMONO POWER AMPLIFIER SERVICE MANUALAlexey OnishenkoNo ratings yet
- Sub-Zero Icing Your Testicles For Increased Male Performance - MyBioHack Unlock Your Maximus PotentialDocument7 pagesSub-Zero Icing Your Testicles For Increased Male Performance - MyBioHack Unlock Your Maximus PotentialfortnitediscordbgweeNo ratings yet
- End Time ProphecyDocument16 pagesEnd Time ProphecyMarven JabianNo ratings yet
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreFrom EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreRating: 4 out of 5 stars4/5 (1090)
- Organizational StrategiesDocument2 pagesOrganizational Strategiesapi-362784633No ratings yet
- SOP - APS - PUR - 02A - Flow Chart For PurchaseDocument2 pagesSOP - APS - PUR - 02A - Flow Chart For Purchaseprakash patelNo ratings yet
- Bonding BB1Document3 pagesBonding BB1DeveshNo ratings yet
- Bi Metallic Corrosion PDFDocument34 pagesBi Metallic Corrosion PDFDerek OngNo ratings yet
- Klubermatic Lubricant DispensersDocument13 pagesKlubermatic Lubricant Dispenserstatankise100% (1)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)From EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Rating: 4.5 out of 5 stars4.5/5 (119)
- ID 2019 CourseDescription MIKE21FlowModelFM HydrodynamicModellingUsingFlexibleMesh UKDocument2 pagesID 2019 CourseDescription MIKE21FlowModelFM HydrodynamicModellingUsingFlexibleMesh UKsaenuddinNo ratings yet
- A300-600 Ammfx1 29 24 00 03 0Document5 pagesA300-600 Ammfx1 29 24 00 03 0Fahimeh HayatinasabNo ratings yet
- Sectors of The Indian EconomyDocument5 pagesSectors of The Indian EconomyhavejsnjNo ratings yet
- State-wise list of 376 approved cold chain projectsDocument45 pagesState-wise list of 376 approved cold chain projectsUsha Hasini VelagapudiNo ratings yet
- Outgassing Properties of Vacuum MaterialsDocument47 pagesOutgassing Properties of Vacuum Materialsmax8086No ratings yet
- Knime SeventechniquesdatadimreductionDocument266 pagesKnime SeventechniquesdatadimreductionramanadkNo ratings yet
- Airworthiness Directives Record ControlDocument4 pagesAirworthiness Directives Record ControlJuan builesNo ratings yet
- Create Windows XP boot CD with McAfee Command Line ScannerDocument3 pagesCreate Windows XP boot CD with McAfee Command Line ScannerSudheesh PuthusseryNo ratings yet
- Problems of Elder AbuseDocument13 pagesProblems of Elder AbuseNeha Jayaraman100% (3)
- Working at Height PolicyDocument7 pagesWorking at Height PolicyAniekan AkpaidiokNo ratings yet
- Wind Energy 6Document12 pagesWind Energy 6Shanthi RameshNo ratings yet
- Sip Annex 3 Gap Analysis Template EditedDocument5 pagesSip Annex 3 Gap Analysis Template EditedRiza GusteNo ratings yet
- Her Body and Other Parties: StoriesFrom EverandHer Body and Other Parties: StoriesRating: 4 out of 5 stars4/5 (821)
- D & C G T G N A. R E S P A, NY 12242: Esign Onstruction Roup HE Overnor Elson Ockefeller Mpire Tate Laza LbanyDocument18 pagesD & C G T G N A. R E S P A, NY 12242: Esign Onstruction Roup HE Overnor Elson Ockefeller Mpire Tate Laza LbanyAlexNo ratings yet
- Welfare Schemes in TelanganaDocument46 pagesWelfare Schemes in TelanganaNare ChallagondlaNo ratings yet