Professional Documents
Culture Documents
Bacterial Antibiotics: Mechanisms of Resistance
Uploaded by
Annisa YohanesOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Bacterial Antibiotics: Mechanisms of Resistance
Uploaded by
Annisa YohanesCopyright:
Available Formats
Mechanisms of Bacterial Resistance to Antibiotics
Laura A. Dever, RPh, Terence S. Dermody, MD
The three fundamental mechanisms of antimicrobial resis- antimicrobial resistance, with emphasis on resistance to anti¬
tance are (1) enzymatic degradation of antibacterial drugs, biotics that inhibit cell-wall biosynthesis. The classification
(2) alteration of bacterial proteins that are antimicrobial targets, system of gram-positive and gram-negative ß-lactamases will
and (3) changes in membrane permeability to antibiotics. Antibi- be detailed, and nonenzymatically mediated ß-lactam resis¬
otic resistance can be either plasmid mediated or maintained on
tance mechanisms will be described. We will also discuss
the bacterial chromosome. The most important mechanism of
resistance to the penicillins and cephalosporins is antibiotic
mechanisms of resistance to the other major classes of antibi¬
hydrolysis mediated by the bacterial enzyme \g=b\-lactamase.The otics, including antagonists of folate synthesis, the aminogly-
expression of chromosomal \g=b\-lactamasecan either be induced cosides, chloramphenicol, and the quinolones.
or stably derepressed by exposure to \g=b\-lactamdrugs. Methods to
RESISTANCE TO ß-LACTAM ANTIBIOTICS
overcome resistance to \g=b\-lactamantibiotics include the develop-
ment of new antibiotics that are stable to \g=b\-lactamaseattack and Penicillins and cephalosporins are the two most commonly
the coadministration of \g=b\-lactamase inhibitors with \g=b\-lactam used classes of ß-lactam antibiotics (Fig 1). Although both
drugs. Resistance to methicillin, which is stable to gram-positive these groups of agents contain a ß-lactam nucleus, they differ
\g=b\-lactamase,occurs through the alteration of an antibiotic target in that penicillins contain a thiazolidine ß-lactam ring complex
protein, penicillin-binding protein 2. Production of antibiotic- and cephalosporins contain a dihydrothiazine ß-lactam ring
modifying enzymes and synthesis of antibiotic-insensitive bac- complex. The antibacterial effect of all ß-lactam antibiotics
terial targets are the primary resistance mechanisms for the
other classes of antibiotics, including trimethoprim, the sulfon- depends on the capacity ofthe antibiotic to diffuse through the
cell membrane, the affinity of the antibiotic for its target
amides, the aminoglycosides, chloramphenicol, and the quino-
lone drugs. Reduced antibiotic penetration is also a resistance proteins, and the stability of the antibiotic against bacterial
mechanism for several classes of antibiotics, including the degradation."
\g=b\-lactamdrugs, the aminoglycosides, chloramphenicol, and the The mechanism ofaction of ß-lactam antibiotics is inhibition
quinolones. of bacterial cell-wall synthesis. Bacterial cell walls are con¬
(Arch Intern Med. 1991;151:886-895) structed from alternating iV-acetylglucosamine and A/-acetyl-
muramic acid residues.5 Transpeptidation is the final step of
cell-wall synthesis and involves the transpeptidase-catalyzed
cross-linking of peptidoglycan chains. As shown in Fig 2,
"Bacterial resistance to antimicrobial agents is becoming transpeptidase-mediated hydrolysis of the ß-lactam bond
increasingly important in clinical practice. While antibiot¬ (CO—N) results in the inhibition of transpeptidation by the
ic resistance is more prevalent in bacteria causing nosocomial covalent linkage of the ß-lactam drug to bacterial transpepti-
infections, the prevalence of antibiotic-resistant pathogenic dase.5 Penicillin-binding proteins (PBPs) are bacterial pro¬
organisms is also increasing in the community at large. ' Forty teins (carboxypeptidases and transpeptidases) responsible
years ago, Staphylococcus aureus was one of the more com¬ for many of the enzymatic activities involved in cell-wall
mon bacterial pathogens (especially in nosocomial infections) synthesis, and they are the primary targets for ß-lactam
and was easily eradicated by penicillin.2,3 At the same time, antibiotics.6 Binding of ß-lactam drugs to PBPs 1, 2, and 3
infections with gram-negative organisms, such as Pseudomo- results in cell-wall lysis, disruption of cell shape, and inhibi¬
nas aeruginosa and Serratia marcescens, were unusual and tion of cell division, respectively.7,8
were often thought to represent colonization rather than ß-Lactam antibiotics can also inhibit bacterial growth by
infection.2 Tbday, most isolates of S aureus are resistant to mechanisms that do not solely involve the inhibition of cell-
penicillin, and infections with gram-negative organisms are wall synthesis. Inhibition of the formation of cell-wall precur¬
common. In addition, many other bacterial pathogens have sors by ß-lactam antibiotics can result in autolysis through the
developed resistance to a large number of antimicrobial unsuppressed activity of murein hydrolases.6 Murein hydro-
agents, and infections with these organisms can be difficult to lases are autolytic enzymes that cause nicks in the cell wall to
treat. provide sites for new peptidoglycan synthesis during cell-wall
Bacteria have developed resistance to every antibiotic used enlargement.9 The inhibition of cell-wall synthesis by ß-lac¬
thus far, and there exist several different mechanisms by tam antibiotics does not alter the activity of these enzymes.10
which antibiotic resistance is mediated (Table). In this re¬ Therefore, bacterial autolysis can result from the effect of
view, we will discuss the most important mechanisms of osmotic pressure on the cell wall damaged by murein hydro¬
Accepted for publication October 16,1990. lases.9,10 In addition to lytic mechanisms, group A streptococci
From the Pharmacy Department, New England Medical Center (Ms Dever); and viridans streptococci do not undergo cell lysis in the
Department of Microbiology and Molecular Genetics, Harvard Medical School presence of ß-lactam drugs (autolysin defective) but can re¬
(Dr Dermody); and Department of Medicine, Brigham and Women's Hospital
(Dr Dermody), Boston, Mass. Dr Dermody is now with the Department of main susceptible to ß-lactam antibiotics by nonlytic mecha¬
Pediatric Infectious Diseases, Vanderbilt University Medical Center, Nash- nisms.11,12
ville, Tenn. The most common cause of resistance to ß-lactam drugs is
Reprint requests to Department ofPediatric Infectious Diseases, Vanderbilt
University Medical Center, Nashville, TN 37232 (Dr Dermody). enzyme-mediated antibiotic degradation. Three classes of
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
enzymes canhydrolyze ß-lactam antibiotics: (1) ß-lactamases, Several different schemes have been used to classify
(2) acylases, and (3) esterases (Fig 3).13 ß-Lactamase-medi- ß-lactamases. Richmond and Sykes identified five classes (I
ated hydrolysis resulting in degradation of the ß-lactam nu¬ through V) of gram-negative ß-lactamases on the basis of
cleus is the most important mechanism of bacterial resistance substrate affinity, isoelectric point, and inhibition data.20 En¬
to ß-lactam antibiotics. ß-Lactamases are produced by some zymes produced by gram-positive organisms (predominantly
gram-positive and all gram-negative bacteria.14,15 ß-Lacta¬ by S aureus) are distinct from those produced by gram-
mases produced by gram-positive organisms are excreted negative organisms. They have a different classification
extracellularly; those produced by gram-negative organisms scheme and are divided into four groups (A through D) pri¬
are usually concentrated in the periplasmic space and not marily by sérologie features.13,21
excreted.15,16 The ß-lactam bond, which is required for the Gram-negative bacilli produce both chromosomally en¬
activity of ß-lactam antibiotics, is also the target of ß-lacta- coded and plasmid-encoded ß-lactamases.18,21 Richmond-
mase attack. ß-Lactamases catalyze the hydrolysis of the Sykes class I and II ß-lactamases, broadly categorized as
ß-lactam bond to acidic derivatives that do not have antibac¬
terial properties (Fig 4).17"19
Genes encoding ß-lactamases can reside on the bacterial
chromosome or on plasmids; bacterial plasmids that encode O
D
proteins responsible for antibiotic resistance are referred to R-C-NH-
as resistance factors (R-factors). Plasmid-mediated resis¬
tance can be passed to distantly related bacterial species by Penicillins
conjugation, and the expression of these enzymes is usually y COOH
constitutive.8 In contrast, the expression of chromosomally
mediated ß-lactamases is usually not constitutive but can be
induced or derepressed by exposure to ß-lactam antibiotics.19 o
II
R-C-NH'
Mechanisms of Bacterial Resistance to Antimicrobial Agents
Cephalosporins
Drug/Drug Class Mechanism of Resistance
Penicillins and Enzymatic inactivation by ß-lactamase; enzymatic
cephalosporins modification by acylase and esterase; outer- COOH
membrane protein deletion; alteration of
penicillin-binding proteins O
Monobactams Enzymatic inactivation by ß-lactamase H
Carbapenems Enzymatic inactivation by ß-lactamase; R-C-NH.
outer-membrane protein deletion
Vancomycin Inhibition of glycopeptide access
Trimethoprim Increased production of dihydrofolate reductase; Monobactams
production of trimethoprim-insensitive Ov •v
dihydrofolate reductase S03H
Sulfonamides Increased production of p-aminobenzoic acid;
increased production of pteridine; increased
production of sulfonamide-insensitive
dihydropteroate synthetase OH
Aminoglycosides Enzymatic modification by acetylation,
phosphorylation, and nucleotidylation;
ribosomal alteration; diminished drug uptake ,NH
Chloramphenicol Enzymatic inactivation by acetylation; decreased Carbapenems •NHCl
drug permeability
Macrolides Enzymatic modification by esterase; alteration of
23S ribosomal RNA
Lincosamides Enzymatic modification by nucleotidylation or
phosphorylation; alteration of 23S ribosomal
RNA
Tetracyclines Active efflux preceded by chemical modification
Quinolones Alteration of subunit A of DNA gyrase; decreased
drug permeability Fig 1.—Structures of ß-lactam antibiotics. The ß-lactam ring (to the left
of the dashed line) is shared by all of these compounds.
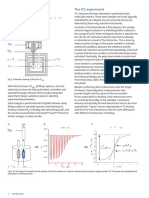
Fig 2.—Inhibition of bacterial transpeptidase by penicillin.
S O
R-C-NH^ ^SV^CH3 Bacterial
>C-
CH3
-;-^
Transpeptidase
R-c-NH^
R C NH ^
N-'s."COOH
s9.
(/ C00H
Transpeptidase
'
II
Penicillin Penicilloyl Enzyme
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
cephalosporinases and penicillinases, respectively, are chro- hyperproduce ß-lactamase despite removal of the inducing
mosomally mediated.20 Induction of class I enzymes occurs ß-lactam antibiotic.24 ß-Lactam drugs that are weak ß-lacta¬
when expression of ß-lactamases increases in the presence of mase inducers can lyse organisms capable of ß-lactamase
a ß-lactam antibiotic (acting as an inducer) and declines to a induction but cannot inhibit bacterial mutants with stably
basal state after hydrolysis or removal of the inducing depressed ß-lactamase expression.22 Weak ß-lactamase in¬
agent.22,23 Citrobacter freundii, Enterobacter cloacae, and ducers used to treat infections caused by P aeruginosa and
P aeruginosa are examples of gram-negative bacteria that E cloacae can lead to ß-lactam resistance through stable
produce class I ß-lactamases that are inducible.24"27 The admin¬ derepression of ß-lactamase expression.22 Cefotaxime, cefti-
istration of strong ß-lactamase inducers, such as cefoxitin and zoxime, and ceftazidime are examples of weak ß-lactamase
imipenem, can lead to the degradation of enzyme-labile ceph¬ inducers capable of selecting stably derepressed mutants.a
alosporins when these drugs are given concomitantly.24,28,29 All of the class III ß-lactamases are plasmid encoded and
Stable derepression of class I ß-lactamase expression can include the TEM-1, TEM-2, and SHV-1 ß-lactamases.20,21 Re¬
occur in bacterial mutants selected for ß-lactam resistance by cently described TEM enzymes mediate the hydrolysis of
exposure to a ß-lactam drug. Stably derepressed mutants aztreonam and the extended-spectrum cephalosporins (eg,
cefotaxime and ceftazidime) and include TEM-3,30,31 TEM-4,32
TEM-7,33 and TEM-10.34 TEM enzymes that hydrolyze ex¬
tended-spectrum cephalosporins usually manifest selective
resistance to specific cephalosporins. Therefore, susceptibil¬
R-C-NH CH3 ity to a particular cephalosporin drug should not be interpret¬
J CH3 ed as representative of an entire cephalosporin class.34 Class
© IV enzymes are chromosomally mediated ß-lactamases pro¬
O
°©—u' Penicillins
COOH
duced by Klebsiella.21 Class V enzymes are a heterogeneous
group of ß-lactamases and include the oxacillin-hydrolyzing
enzymes of Enterobacteriaceae (OXA-1, OXA-2, and OXA-3)
and the Pseudomonas-speciftc enzymes.20,21
Resistance to ß-lactam antibiotics can be inhibited through
II the chemical modification of ß-lactam drugs to improve stabil¬
R-C-NH-
ity against ß-lactamase hydrolysis. Aztreonam and imi-
o penem, each representing a unique ß-lactam antibiotic class,
© CH2-0 —
II
C-CH3
are two of the most promising new agents (Fig l).35,36 Az¬
treonam is a monobactam that is active only against gram-
w
negative aerobic organisms.37,38 It demonstrates an especially
COOH © high degree of stability to plasmid-encoded ß-lactamases38
and does not induce chromosomally encoded ß-lactamases.37
Cephalosporins Imipenem (A^-formimidoyl thienamycin) is a carbapenem and
is similar to aztreonam in that it is highly resistant to
Fig 3.—Sites for enzymatic degradation of penicillins and cephalospo¬ ß-lactamase degradation.36,39,40 Imipenem is degraded by renal
rins: ß-lactamase (1 ), acylase (2), and esterase (3). dihydropeptidase and must be coadministered with cilastatin,
Fig 4.—Reactions catalyzed by ß-lactamases resulting in the degradation of penicillins (top) and
cephalosporins (bottom).
O O
II II
R-C-NH R-C- NH
:.-«: ¿f=C Nc CH3
CH3
CH3 CH3
COOH O OH
| COOH
I_I
L_H__!
COOH
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
a dihydropeptidase inhibitor. Imipenem is active against a
large number of gram-positive, gram-negative, and anaerobic CHjOH
organisms, including Bacteroides fragilis, Escherichia coli,
P aeruginosa, and S aureus.41*1 It is not affected by a perme¬
ability barrier and can readily enter into the periplasm. Al¬ \*
though imipenem is stable against most plasmid-encoded and
chromosomally encoded ß-lactamases,15,36 zinc-containing ß-
lactamases that are capable of imipenem hydrolysis have been
identified in strains of Xanthomonas maltophilia44 and "COO-K*
B fragilis.45
The coadministration of a nonantimicrobial drug capable of Clavulanate Potassium
inhibiting ß-lactamase activity in conjunction with a ß-lactam
antibiotic is a strategy that has recently been used to over¬
come ß-lactamase-mediated resistance. Clavulanate and sul-
bactam, two prototype agents now in widespread use, both
act irreversibly to inhibit ß-lactamase activity4,14 (Fig 5). The
combination of either of these agents with ampicillin results in
antibacterial activity against ampicillin-resistant strains of
Bacteroides, E coli, Haemophilus influenzae, and S aur-
ews.4,46
Hydrolysis of clavulanate is followed by (1) formation of a •COO"Na+
long-lived acyl-enzyme intermediate, (2) transient regenera¬
tion of free ß-lactamase, and then (3) irreversible ß-lactamase Sulbactam Sodium
inactivation through the formation of a covalent linkage be¬
tween the enzyme active site and the inhibitor.47,48 Hydrolysis Fig 5.—Structures of clavulanate potassium and sulbactam sodium.
of sulbactam results in the formation of sulbactam fragments Both compounds are noncompetitive ß-lactamase inhibitors.
that covalently bond to a site on the enzyme distinct from the
active site, causing irreversible enzyme inactivation.13 Clavu¬ strains that express PBP 2a constitutively.56,61 Ordinarily,
lanate and sulbactam can inhibit plasmid-encoded ß-lacta¬ within a given S aureus culture, few of the isolates express
mases (Richmond-Sykes classes III and V) and chromosomal¬ methicillin resistance (heterogeneous resistance),62 but occa¬
ly encoded ß-lactamases (Richmond-Sykes classes II and IV) sionally all of the isolates are resistant (homogeneous resis¬
of gram-negative bacteria and ß-lactamases of gram-positive tance).63 The level of methicillin resistance in culture does not
bacteria.46 Clavulanate and sulbactam are poor inhibitors of have clinical significance in that all of these isolates should be
class I enzymes4*"1;clavulanate induces these enzymes in considered resistant to methicillin.
some bacteria, especially P aeruginosa and Enterobacter.m A novel resistance mechanism has recently been described
Because of its capacity to induce ß-lactamase production, in methicillin-resistant S aureus with low or borderline levels
clavulanate may antagonize the effect of ticarcillin on strains of resistance to ß-lactam antibiotics. These methicillin-resis¬
of Morganella morganii and E cloacae that are capable of tant strains do not express PBP 2a64,65 but possess PBPs that
induced ß-lactamase expression.50 In contrast to clavulanate, have modified penicillin-binding capacities (particularly PBP
sulbactam is a less potent inhibitor of most ß-lactamases. 1 and PBP 2).ffi Compared with methicillin-resistant S aureus
However, sulbactam is a poor ß-lactamase inducer and thus strains that produce PBP 2a, the minimum inhibitory concen¬
offers a distinct advantage over clavulanate.52 trations of methicillin for methicillin-resistant S aureus with
RESISTANCE TO METHICILLIN modified penicillin-binding capacities are low, and therefore
these organisms generally remain susceptible to methicillin.
Alteration ofthe ß-lactam nucleus has yielded many antimi¬
However, this mechanism of resistance could be an important
crobials (eg, methicillin, nafcillin, and oxacillin) with in¬ cause of methicillin resistance in the future.65
creased stability to gram-positive ß-lactamases. Resistance
to these agents does occur, however, and is independent of RESISTANCE TO ß-LACTAM ANTIBIOTICS
THROUGH REDUCED PENETRATION
ß-lactamase production. Methicillin-resistant strains of
S aureus are increasingly common in nosocomial53 infections Porin proteins in the outer membrane of gram-negative
and have also been described in infections of intravenous- bacteria form channels that allow the exchange of nutrients
drug abusers.54,55 Methicillin resistance is mediated through and other substances (including antibiotics) between the ex¬
the production of an altered PBP 2 (PBP 2a or PBP 2')f*58 that tracellular environment and the periplasmic space.66"68 For
has a much lower affinity for methicillin (and for most other ß-lactam antibiotics to bind PBPs, they must penetrate porin
ß-lactam antibiotics) than does PBP 2.59 channels in the outer membrane and traverse the periplasmic
Methicillin-resistant S aureus can exhibit either high or low space.69 Diffusion of some ß-lactam drugs through porin chan¬
levels of resistance. Whereas greater amounts of PBP 2a are nels is limited by the presence of bulky substituents on the
produced in high-level methicillin-resistant S aureus, there is acyl side chain of the ß-lactam nucleus; diffusion of others is
no correlation between PBP 2a production and minimum limited by electrostatic charge.70 Changes in outer membrane
inhibitory concentration.60 The expression of PBP 2a can ei¬ permeability serve to limit drug influx and decrease drug
ther be induced by methicillin (or other ß-lactam antibiotics) availability to PBPs located on the inner membrane.23,68
or be constitutive. A ß-lactamase plasmid is usually present in Two outer membrane proteins (Omps) of E coli that have
inducible methicillin-resistant S aureus strains but absent in been identified and characterized are OmpF and OmpC.66,70,71
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
OH
\ HO
H>C \ . \ \ç_— CH2OH
H°CH3 \^°
CI
h0y-^/^ci O
%X
O
^/\/oh O CH3
11 H X H m
II I
I
H
h
CH2
i! H
TCH2
HN\\ h
A I
c=0
I
CH
HO / \ OH
HO
Fig 6.—Structure of vancomycin showing the site of the A/-substitution. R =
H in vancomycin.
An additional (mutant) channel, PhoE, is produced in mu¬ RESISTANCE TO VANCOMYCIN
tants of E coli deficient in OmpF and OmpC under conditions Vancomycin is a glycopeptide antibiotic (Fig 6) that inhibits
of phosphate depletion.66 Of these three porin channels, bacterial cell-wall formation by the inhibition of peptidogly¬
OmpF has the widest pore diameter and the highest perme¬ can synthesis. It binds to the free carboxyl end of D-alanyl-D-
ability to ß-lactam drugs.71 Zwitterionic compounds (eg, am- alanine of V-acetyl-muramyl-pentapeptide of the elongating
picillin, cefaclor, cephalexin, and imipenem) have much high¬ peptidoglycan chain.77,78 Vancomycin activity is limited to
er penetration rates through OmpF than do anionic
gram-positive organisms, and the drug is especially useful in
compounds (eg, carbenicillin, cefamandole, cefuroxime, and the treatment of infections caused by methicillin-resistant
piperacillin).70 Permeability of zwitterionic compounds S aureus and penicillin-resistant strains of Enterococcus.™
through PhoE channels is poor; in contrast, anionic com¬ Resistance to vancomycin has recently been observed in
pounds have higher penetration rates through PhoE channels Enterococcus faecium60^2 and Enterococcus faecalis.® In
than through OmpF channels.71 Escherichia coli mutants de¬ these organisms, vancomycin resistance is plasmid mediated
ficient in OmpF channels are resistant to ß-lactam antibiotics and inducible.82^4 Although the mechanism of resistance is
as a consequence of reduced penetration through OmpC chan¬
poorly understood, novel cytoplasmic membrane proteins
nels produced by these organisms.70"72 In addition, enzymatic have been identified in vancomycin-resistant strains of
degradation of ß-lactam antibiotics is more efficient in OmpF- Efaecium'1 and Efaecalis.^ It has been suggested that these
deficient mutants because of the slower drug penetration that
proteins block vancomycin access to the elongating peptido¬
occurs through OmpC channels.68 glycan chain.81,83 Strains of Efaecium and Efaecalis that are
Penetration of compounds through the outer membrane of resistant to vancomycin maintain susceptibility to some N-
P aeruginosa is less than that of E coli, primarily because of substituted vancomycin derivatives (eg, decyl-vancomycin,
the low permeability of its porin channels.67 Impaired pene¬ p-octyloxybenzyl-vancomycin)
tration appears to be the mechanism of imipenem resistance
p-octylbenzyl-vancomycin,
exhibited by some strains of P aeruginosa.13 Loss of 45- to
(Figo).84
48-kd Omps has been reported in strains of P aeruginosa that RESISTANCE TO THE FOLATE ANTAGONISTS:
TRIMETHOPRIM AND THE SULFONAMIDES
are imipenem resistant.73"75 These proteins correspond to
porin channel D2 which is required for the penetration of Trimethoprim and the sulfonamides act synergistically to
imipenem.74 inhibit folate metabolism by blocking sequential steps in the
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
Most aminoglycosides (amikacin, gentamicin, neomycin, ne-
p-Aminobenzoic Acid + Pteridine tilmicin, and tobramycin) contain 2-deoxystreptamine; strep¬
tomycin contains streptidine. Aminoglycosides are hydro-
Dihydropteroate philic molecules that require active transport to gain entry
Synthetase into bacteria.96 The primary mechanism of aminoglycoside
resistance is enzyme-mediated antibiotic modification. Alter¬
Sulfonamides h-** ation ofribosomal binding sites96 and diminished drug uptake98
are less common mechanisms of aminoglycoside resistance.
S02NH Dihydropteroic Acid Aminoglycoside-modifying enzymes (AMEs) catalyze the
modification of either the amino or hydroxyl groups on amino-
T~\ Dihydrofolate
Synthetase
cyclitol and aminoglycoside antibiotics (Fig S).m,S9 These en¬
zymes inactivate aminoglycosides by three mechanisms: (1)
NH2
acetylation, (2) nucleotidylation, and (3) phosphorylation.99"102
Bacterial AMEs of most gram-positive and gram-negative
Sulfamethoxazole Dihydrofolic Acid organisms are plasmid encoded.97 Concentrations of AMEs in
the periplasmic space of gram-negative bacteria are usually
Dihydrofolate low.101 However, modification of aminoglycosides is efficient
Reductase
because antibiotic uptake into the periplasmic space is slow,
and AME affinity for aminoglycosides is high.101 Each AME
Trimethoprim r—** inactivates a particular subset of the available aminoglycoside
antibiotics accounting for the varied spectra of aminoglyco¬
side resistance observed in clinical isolates.101
OCH3 Tetrahydrofolic Acid
Aminoglycoside acylases inactivate aminoglycosides by
OCH3 catalyzing the transfer of acetate from acetyl-coenzyme A to
an amino group on 2-deoxystreptamine-containing aminogly¬
OCH3 cosides, such as amikacin, gentamicin, and neomycin.97,101
Aminoglycoside nucleotidyltransferases utilize adenosine tri-
Fig 7.—Sites of inhibition of folate metabolism by sulfonamides and phosphate and other nucleotide triphosphates as cofactors
trimethoprim. The action of these agents on two independent bio¬ and adenylylate hydroxyl groups on amikacin, gentamicin,
chemical steps is responsible for the synergy observed with these
antibiotics. kanamycin, neomycin, spectinomycin, streptomycin, and to¬
bramycin.97 Aminoglycoside phosphotransferases catalyze
the transfer of a phosphate group to a hydroxyl group on
amikacin, gentamicin, kanamycin, neomycin, and tobramy¬
folate synthesis pathway (Fig 7). Trimethoprim inhibits dihy¬ cin. 1M A single aminoglycoside can be altered at several differ¬
drofolate reductase, and the sulfonamides inhibit dihydro¬ ent sites by the coupling of one or more AMEs, a process
pteroate synthetase.85,86 Bacteria are not able to take up pre¬ resulting in the complete inactivation of the antibiotic.97100
formed folie acid derivatives and cannot utilize host folie acid. Recently, strains of E faecalis and E faecium have been
Therefore, bacterial production of tetrahydrofolic acid, which reported that show high levels of aminoglycoside resistance
is required for the synthesis of amino acids and nucleotides, is (minimum inhibitory concentration, >2000 mg/L).103 Effec¬
entirely dependent on synthesis from pteridine and p-amino- tive treatment of serious infections caused by these organ¬
benzoicacid.85 isms requires the synergistic combination of ampicillin, peni¬
The major cause of trimethoprim resistance is the produc¬ cillin, or vancomycin and an aminoglycoside.104"106 Enterococci
tion of an altered (plasmid-encoded) dihydrofolate reductase with high levels of aminoglycoside resistance can be especial¬
that lacks the capacity to bind trimethoprim.87'91 Overproduc¬ ly difficult to treat, as therapy with a cell-wall synthesis
tion of the normal (chromosomally encoded) dihydrofolate inhibitor alone is usually not effective. Furthermore, entero¬
reductase is an additional mechanism of trimethoprim resis¬ cocci that manifest high levels of aminoglycoside resistance
tance, particularly in E coli and Klebsiella pneumoniaef,SZlXI can exhibit resistance to several aminoglycosides simulta¬
Bacterial resistance to the sulfonamides may be caused by neously.103107 However, individual aminoglycoside susceptibil¬
increased production of p-aminobenzoic acid, increased syn¬ ity testing is advisable, since enterococci can demonstrate
thesis of pteridine, and increased production of a sulfonamide- high levels of aminoglycoside resistance to gentamicin, kana¬
resistant dihydropteroate synthetase.87,94 Most resistance to mycin, tobramycin, and streptomycin despite only moderate
sulfonamides, however, is due to an additional plasmid-en¬ resistance to amikacin or netilmicin.103,108
coded dihydropteroate synthetase that has a lowered affinity RESISTANCE TO CHLORAMPHENICOL
for sulfonamides.85,87,95 As is the case with ß-lactam antibiotics,
loss of inhibitory activity ofeither trimethoprim or the sulfon¬ Chloramphenicol inhibits bacterial protein synthesis by
amides can result from impaired penetration.87,98 binding to the 50S ribosomal subunit.109 The most common
mechanism of resistance to chloramphenicol is enzymatic
RESISTANCE TO AMINOGLYCOSIDES
modification.110,111 Chloramphenicol acetyltransferase is an in-
Aminoglycoside antibiotics inhibit the initiation of protein ducible, plasmid-encoded enzyme that inactivates chloram¬
synthesis and disrupt polypeptide chain elongation by induc¬ phenicol by acetylation (Fig 9).112"114 Decreased chlorampheni¬
ing the incorporation of incorrect amino acids.96 Aminoglyco- col permeability has also been proposed to be a cause of
sides are classified into two groups, depending on whether resistance in H influenzaem and Pseudomonas cepacia.116 In
they contain 2-deoxystreptamine or streptidine (Fig 8).97 chloramphenicol-resistant strains of H influenzae that do not
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
C. D- AAC m
NH AAC (6')
II
NHCNH2 NH CH2NH2 CH2NH2
O/TON™"™2 APH (3')
(41)—MHOVH A
APH (3')
(4')—K3H^H /
}- OAjj)
V_/^APH (6)
~ANT(6)
ANT
AAC (2')
ANT
AAC (2') —* H2N
O
AAC (3)-AhT\^ AAC (3) -^hT\^
NH2
OH OH NH2
APH (5") —*HOÇH2 0 o
CH2OH
ANT (3") hoNíiv
ANT (2") —»OH
APH (3") APH (2")
Fig 8.—Aminoglycoside structures and sites for enzymatic modification. A, Spectinomycin; B, streptomy¬
cin; C, neomycin; and D, kanamycin B. Amikacin, gentamicin, and tobramycin are structurally related to
kanamycin B. AAC indicates aminoglycoside A/-acetyltransferase; AAD, aminoglycoside adenylyltrans-
ferase; ANT, aminoglycoside nucleotidyltransferase; and APH, aminoglycoside phosphotransferase
(modified from Phillips and Shannon97).
C-CHCI2 C-CHCI2
Chloramphenicol
OH
NH
Acetyltransferase VC
C—C- CH2OH OM-C —
C —
CH20 —
COCH3
OH H OH H
Chloramphenicol 3-O-Acetyl Chloramphenicol
Acetyl CoA
Fig 9.—Acetylation of chloramphenicol by chloramphenicol acetyltransferase. CoA indicates coenzyme
A.
produce chloramphenicol acetyltransferase, reduced quanti¬ matic modification, does not confer resistance to this
ties of an Omp have been observed.115 It has been suggested drug.124,125
that the loss of this Omp in resistant strains results in de¬ RESISTANCE TO QUINOLONES
creased chloramphenicol permeability.115
MECHANISMS OF RESISTANCE TO OTHER
The 6-fluoroquinolone antibiotics, ciprofloxacin and nor-
CLASSES OF ANTIBIOTICS
floxacin, were derived from nalidixic acid.126 These drugs
exhibit bactericidal activity against a wide range of gram-
Macrolide (erythromycin) and lincosamide (clindamycin) positive and gram-negative bacteria, including some strains
antibiotics inhibit bacterial protein synthesis by binding to of methicillin-resistant S aureus and P aeruginosa
the 50S ribosomal subunit.117 Resistance to these agents is (Fig 10).127"130 Quinolone antibiotics inhibit DNA gyrase, an
mediated by mechanisms similar to those responsible for enzyme required for unwinding of the supercoiled bacterial
aminoglycoside resistance. Macrolide and lincosamide resis¬ chromosome before DNA replication.131 DNA gyrase is com¬
tance is due to either alteration of the ribosomal target site posed of two A subunits (encoded by gyrA) and two B sub-
(adenine methylation in 23S ribosomal RNA)118 or enzyme- units (encoded by gyrB).1S2,133
mediated antibiotic modification.119 Resistance to nalidixic acid and the 6-fluoroquinolone drugs
The tetracyclines act to inhibit bacterial protein synthesis occurs through two independent mechanisms, and neither
by binding to the 30S ribosomal subunit.120 Resistance to mechanism is plasmid mediated.132 Although resistance to
tetracycline is also caused by enzymatic modification,121 and nalidixic acid is mediated by the same mechanisms as resis¬
alteration of this drug is followed by rapid efflux from the tance to the 6-fluoroquinolone antibiotics,134 resistance to nali¬
cytoplasm.122,123 Tetracycline efflux, unaccompanied by enzy- dixic acid is 100- to 1000-fold greater than resistance to the
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
CONCLUSION
o As discussed in this review, bacteria have developed resis¬
II tance to all classes of antimicrobial agents curently in use.
.COOH Furthermore, newer mechanisms of antibiotic resistance
have caused significant problems in the treatment of infec¬
tions caused by some bacteria, such as enterococci and Pseu-
domonas. New strategies to combat antibiotic resistance
H.C^S, include the modification of antibiotics now in use and the
coadministration of nonantibiotic drugs that can inhibit anti¬
I biotic degradation mediated by bacterial enzymes. Despite
C2H5
these technological innovations, it is very likely that antibiot¬
Nalidixic Acid ic resistance will continue to be an important clinical problem.
Several methods have been developed to decrease the risk of
infection with antibiotic-resistant organisms. These include
choosing an antibiotic with a narrow spectrum when a patho¬
gen is known, the use of short durations of antibiotic prophy¬
COOH laxis, and limiting topical and oral therapy with drugs that
may have to be used parenterally. While it remains controver¬
sial whether the indiscriminate use of antibiotics actually
increases the emergence of drug-resistant pathogens, it
seems prudent to use these drugs carefully to prolong their
efficacy.
This study was supported in part by Physician-Scientist Award 5K11
AI00865 from the National Institute of Allergy and Infectious Diseases, Be-
Norfloxacin thesda, Md (Dr Dermody).
We express our appreciation to Jerry Jones, PhD, James Maguire, MD,
Thomas O'Brien, MD, and Richard T. Scheife, PharmD, for essential discus¬
sions and reviews of the manuscript.
References
1. Young LS. Antimicrobial resistance: implications for antibiotic use. Am J
COOH Med. 1986;80(suppl 5C):35-39.
2. Weinstein L. Gram-negative bacterial infections: a look at the past, a view
of the present, and a glance at the future. Rev Infect Dis. 1985;7(suppl 4):538\x=req-\
544.
3. Murray BE. Problems and mechanisms of antimicrobial resistance. Infect
Dis Clin North Am. 1989;3:423-439.
4. Campoli-Richards DM, Brogden RN. Sulbactam/ampicillin: a review of
its antibacterial activity, pharmacokinetic properties, and therapeutic use.
Drugs. 1987;33:577-609.
5. Blumberg PM, Strominger JL. Interaction of penicillin with the bacterial
cell: penicillin-binding proteins and penicillin-sensitive enzymes. Bacteriol
Rev. 1974;38:291-335.
Ciprofloxacin 6. Tomasz A. The mechanism of the irreversible antimicrobial effects of
penicillins:how the beta-lactam antibiotics kill and lyse bacteria. Ann Rev
Microbiol. 1979;33:113-137.
Fig 10.—Structures of nalidixic acid and the 6-fluoroquinolone com¬ 7. Spratt BG. Distinct penicillin binding proteins involved in the division,
pounds norfloxacin and ciprofloxacin. elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A.
1975;72:2999-3003.
8. Waxman DJ, Strominger JL. Penicillin-binding proteins and the mecha-
6-fluoroquinolones.135 The most common mechanism of resis¬ nism of action of \g=b\-lactamantibiotics. Ann Rev Biochem. 1983;52:825-869.
tance to quinolone antibiotics is alteration of the target en¬ 9. Higgins ML, Shockman GD. Procaryotic cell division with respect to wall
and membranes. CRC Crit Rev Microbiol. 1971;1:29-72.
zyme. The A subunit of DNA gyrase is altered in quinolone- 10. Weidel W, Pelzer H. Bagshaped macromolecules\p=m-\a new outlook on
resistant gyrA mutants so that it no longer binds the bacterial cell walls. Adv Enzymol. 1964;26:193-232.
antibiotic.136"138 Quinolone resistance also occurs through de¬ 11. Gutmann L, Williamson R, Tomasz A. Physiological properties of peni-
creased penetration.132 Quinolone antibiotics rapidly pene¬ cillin-binding proteins group streptococci. Antimicrob Agents Chemother.
in A
1981;19:872-880.
trate the outer membrane of susceptible strains of E coli 12. Horne D, Tomasz A. Tolerant response of Streptococcus sanguis to beta\x=req-\
lactams and other cell wall inhibitors. Antimicrob Agents Chemother.
through OmpF channels.139 Mutations in regulatory loci se¬ 1977;11:888-896.
lected for ciprofloxacin (cfxB) and norfloxacin (nfxB) resis¬ 13. Noguchi JK, Gill MA. Sulbactam: a \g=b\-lactamaseinhibitor. Clin Pharm.
tance, respectively, are associated with diminished drug pen¬ 1988;7:37-51.
14. Neu HC, Fu KP. Clavulanic acid, a novel inhibitor of \g=b\-lactamases.
etration through decreased expression of OmpF.132,135,140 Antimicrob Agents Chemother. 1978;14:650-655.
Alterations in outer membrane permeability have also been 15. Neu HC. Carbapenems: special properties contributing to their activity.
Am J Med. 1985;78(suppl 6A):33-40.
reported in quinolone-resistant strains of P aeruginosa. The 16. Nord CE. Mechanisms of \g=b\-lactamresistance in anaerobic bacteria. Rev
precise mechanism of decreased quinolone permeability is not Infect Dis. 1986;8(suppl 5):543-548.
known, but it cannot be attributed to a decrease in OmpF 17. Fisher J, Belasco JG, Charnas RL, et al. \g=b\-Lactamaseinactivation by
mechanism-based reagents. Phil Trans R Soc Lond Biol. 1980;289(ser B):309\x=req-\
expression.141,142 Quinolone antibiotics can be suitable alterna¬ 319.
tive agents in the treatment of some strains of methicillin- 18. Sykes RB, Matthew M. The \g=b\-lactamasesof gram-negative bacteria and
resistant S aureus; however, high-level quinolone resistance their role in resistance to \g=b\-lactam antibiotics. J Antimicrob Chemother.
has recently been reported in strains of S aureus that are also 1976;2:115-157.
19. Sanders CC. Inducible \g=b\-lactamases and non-hydrolytic resistance
highly resistant to gentamicin143 and methicillin.143,144 mechanisms. J Antimicrob Chemother. 1984;13:1-3.
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
20. Richmond HM, Sykes RB. The \g=b\-lactamasesof gram-negative bacilli and 568.
their possible physiologic role. Adv Microbiol Physiol. 1973;9:31-88. 51. Hunter PA, Coleman K, Fisher J, et al. In vitro synergistic properties of
21. Sykes RB. The classification and terminology of enzymes that hydrolyze clavulanic acid, with ampicillin, amoxycillin and ticarcillin. J Antimicrob Che-
\g=b\-lactamantibiotics. J Infect Dis. 1982;145:762-765. mother. 1980;6:455-470.
22. Livermore DM. Clinical significance of beta-lactamase induction and 52. Labia R, Morand A, Lelievre V, et al. Sulbactam: biochemical factors
stable derepression in gram-negative rods. Eur J Clin Microbiol. 1987;6:439\x=req-\ involved in its synergy with ampicillin. Rev Infect Dis. 1986;8(suppl 5):496-502.
445. 53. Hunt JL, Purdue GF, Tuggle DW. Morbidity and mortality of an endem-
23. Sanders CC, Sanders WE. Clinical importance of inducible beta-lacta- ic pathogen: methicillin-resistant Staphylococcus aureus. Am J Surg.
mases in gram-negative bacteria. Eur J Clin Microbiol. 1987;6:435-438. 1988;156:524-528.
24. Shannon K, Phillips I. The effects on \g=b\-lactamsusceptibility ofphenotyp- 54. Crane LR, Levine DP, Zervos MJ, et al. Bacteremia in narcotic addicts
ic induction and genotypic derepression of \g=b\-lactamasesynthesis. J Antimicrob at the Detroit Medical Center, I: microbiology, epidemiology, risk factors, and
Chemother. 1986;18(suppl E):15-22. empiric therapy. Rev Infect Dis. 1986;8:364-373.
25. Livermore DM, Akova M, Wu P, et al. Clavulanate and \g=b\-lactamase 55. Craven DE, Rixinger AI, Goularte TA, et al. Methicillin-resistant
induction. J Antimicrob Chemother. 1989;24(suppl B):23-33. Staphylococcus aureus bacteremia linked to intravenous drug abusers using a
26. Sanders CC, Sanders WE. Emergence of resistance during therapy with 'shooting gallery.' Am J Med. 1986;80:770-776.
the newer \g=b\-lactamantibiotics: role of inducible \g=b\-lactamasesand implications 56. Ubukata K, Yamashita N, Konno M. Occurrence of a \g=b\-lactam-inducible
for the future. Rev Infect Dis. 1983;5:639-648. penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob
27. Vu H, Nikaido H. Role of \g=b\-lactamhydrolysis in the mechanism of Agents Chemother. 1985;27:851-857.
resistance of a \g=m\-lactamase-constitutiveEnterobacter cloacae strain to ex- 57. Utsui Y, Yokata T. Role of an altered penicillin-binding protein in
panded-spectrum \g=b\-lactams.Antimicrob Agents Chemother. 1985;27:393-398. methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents
28. Sanders CC, Sanders WE. Type I \g=b\-lactamasesof gram-negative bacte- Chemother. 1985;28:397-403.
ria: interactions with \g=b\-lactamantibiotics. J Infect Dis. 1986;154:792-800. 58. Murakami K, Tomasz A. Involvement of multiple genetic determinants
29. Cullmann W, Buscher KH, Dick W. Selection and properties of Pseudo- in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol.
monas aeruginosa variants resistant to beta-lactam antibiotics. Eur J Clin 1989;171:874-879.
Microbiol. 1987;6:467-473. 59. Hartman BJ, Tomasz A. Low-affinity penicillin-binding protein associat-
30. Kitzis MD, Billot-Klein D, Goldstein FW, et al. Dissemination of the ed with \g=b\-lactam resistance in Staphylococcus aureus. J Bacteriol.
novel plasmid-mediated \g=b\-lactamaseCTX-1, which confers resistance to broad- 1984;158:513-516.
spectrum cephalosporins, and its inhibition by \g=b\-lactamaseinhibitors. Antimi- 60. Murakami K, Nomura K, Doi M, et al. Production of low-affinity penicil-
crob Agents Chemother. 1988;32:9-14. lin-binding protein by low- and high-resistance groups of methicillin-resistant
31. Petit A, Sirot DL, Chanal CM, et al. Novel plasmid-mediated \g=b\-lacta- Staphylococcus aureus. Antimicrob Agents Chemother. 1987;31:1307-1311.
mase in clinical isolates of Klebsiella pneumoniae more resistant to ceftazidime 61. Ubukata K, Nonoguchi R, Matsuhashi M, et al. Expression and induc-
than to other broad-spectrum cephalosporins. Antimicrob Agents Chemother. ibility in Staphylococcus aureus ofthe mecA gene, which encodes a methicillin\x=req-\
1988;32:626-630. resistant S. aureus\p=m-\specificpenicillin-binding protein. J Bacteriol.
32. Paul GC, Gerbaud G, Bure A, et al. TEM-4, a new plasmid-mediated \g=b\ x=req-\ 1989;171:2882-2885.
lactamase that hydrolyzes broad-spectrum cephalosporins in a clinical isolate of 62. Hartman BJ, Tomasz A. Expression of methicillin resistance in hetero-
Escherichia coli. Antimicrob Agents Chemother. 1989;33:1958-1963. geneous strains of Staphylococcus aureus. Antimicrob Agents Chemother.
33. Gutmann L, Kitzis MD, Billot-Klein D, et al. Plasmid-mediated \g=b\ x=req-\ 1986;29:85-92.
lactamase (TEM-7) involved in resistance to ceftazidime and aztreonam. Rev 63. Kornblum J, Hartman BJ, Novick RP, et al. Conversion of a homoge-
Infect Dis. 1988;10:860-865. neously methicillin-resistant strain of Staphylococcus aureus to heterogeneous
34. Quinn JP, Miyashiro D, Sahm D, et al. Novel plasmid-mediated \g=b\ x=req-\ resistance by Tn551-mediated insertional inactivation. Eur J Clin Microbiol.
lactamase (TEM-10) conferring selective resistance to ceftazidime and az- 1986;5:714-718.
treonam in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents 64. Chambers HF, Archer G, Matsuhashi M. Low-level methicillin resis-
Chemother. 1989;33:1451-1456. tance in strains of Staphylococcus aureus. Antimicrob Agents Chemother.
35. Bush K, Freudenberger JS, Sykes RB. Interaction of azthreonam and 1989;33:424-428.
related monobactams with \g=b\-lactamasesfrom gram-negative bacteria. Antimi- 65. Tomasz A, Drugeon HB, De Lencastre HM, et al. New mechanism of
crob Agents Chemother. 1982;22:414-420. methicillin resistance in Staphylococcus aureus: clinical isolates that lack the
36. Richmond MH. The semi-synthetic thienamycin derivative MK0787 and PBP 2a gene and contain normal-penicillin-binding proteins with modified
its properties with respect to a range of \g=b\-lactamasesfrom clinically relevant penicillin-binding capacity. Antimicrob Agents Chemother. 1989;33:1869-1874.
bacterial species. J Antimicrob Chemother. 1981;7:279-285. 66. Nikaido H, Rosenberg EY, Foulds J. Porin channels in Escherichia coli.
37. Sykes RB, Bonner DP. Aztreonam: the first monobactam. Am J Med. J Bacteriol. 1983;153:232-240.
1985;78(suppl 2A):2-10. 67. Yoshimura F, Zalman LS, Nikaido H. Purification and properties of
38. Georgopapadakou NH, Smith SA, Sykes RB. Mode of action of azth- Pseudomonas aeruginosa porin. J Biol Chem. 1983;258:2308-2314.
reonam. Antimicrob Agents Chemother. 1982;21:950-956. 68. Nikaido H. Bacterial resistance to antibiotics as a function of outer
39. Acar JF, Goldstein FW, Kitzis MD, et al. Activity of imipenem on membrane permeability. J Antimicrob Chemother. 1988;22(suppl A):17-22.
aerobic bacteria. J Antimicrob Chemother. 1983;12(suppl D):37-45. 69. Sanders CC. Novel resistance selected by the new expanded-spectrum
40. Neu HC, Labthavikul P. Comparative in vitro activity of N-formimidoyl cephalosporins: a concern. J Infect Dis. 1983;147:585-589.
thienamycin against gram-positive and gram-negative aerobic and anaerobic 70. Yoshimura F, Nikaido H. Diffusion of \g=b\-lactamantibiotics through the
species and its \g=b\-lactamase stability. Antimicrob Agents Chemother. porin channels of Escherichia coli K-12. Antimicrob Agents Chemother.
1982;21:180-187. 1985;27:84-92.
41. Tally FP, Jacobus NV. Susceptibility of anaerobic bacteria to imipenem. 71. Nikaido H, Rosenberg EY. Porin channels in Escherichia coli: studies
J Antimicrob Chemother. 1983;12(suppl D):47-51. with liposomes reconstituted from purified proteins. J Bacteriol. 1983;153:241\x=req-\
42. Kesado T, Hashizume T, Asahi Y. Antibacterial activities of a new 252.
stabilized thienamycin, N-formimidoyl thienamycin, in comparison with other 72. Harder KJ, Nikaido H, Matsuhashi M. Mutants of Escherichia coli that
antibiotics. Antimicrob Agents Chemother. 1980;17:912-917. are resistant to certain beta-lactam compounds lack the ompF porin. Antimi-
43. Mitsuhashi S. In vitro and in vivo antibacterial activity of imipenem crob Agents Chemother. 1981;20:549-552.
against clinical isolates of bacteria. J Antimicrob Chemother. 1983;12(suppl 73. Buscher KH, Cullmann W, Dick W, et al. Imipenem resistance in Pseu-
D):53-64. domonas aeruginosa resulting from diminished expression of an outer mem-
44. Saino Y, Kobayashi F, Inoue M, et al. Purification and properties of brane protein. Antimicrob Agents Chemother. 1987;31:703-708.
inducible penicillin \g=b\-lactamaseisolated from Pseudomonas maltophilia. Anti- 74. Lynch MJ, Drusano GL, Mobley HLT. Emergence of resistance to
microb Agents Chemother. 1982;22:564-570. imipenem in Pseudomonas aeruginosa. Antimicrob Agents Chemother.
45. Cuchural GJ, Malamy MH, Tally FP. \g=b\-Lactamase\p=m-\mediatedimipenem 1987;31:1892-1896.
resistance in Bacteroides fragilis. Antimicrob Agents Chemother. 1986;30:645\x=req-\ 75. Quinn JP, Dudek EJ, DiVincenzo CA, et al. Emergence of resistance to
648. imipenem during therapy for Pseudomonas aeruginosa infections. J Infect
46. Aldridge KE, Sanders CV, Marier RL. Variation in the potentiation of Dis. 1986;154:290-294.
\g=b\-lactamantibiotic activity by clavulanic acid and sulbactam against multiply 76. Trias J, Nikaido H. Outer membrane protein D2 catalyzes facilitated
antibiotic-resistant bacteria. J Antimicrob Chemother. 1986;17:463-469. diffusion of carbapenems and penems through the outer membrane of Pseudo-
47. Fisher J, Charnas RL, Knowles JR. Kinetic studies on the inactivation of monas aeruginosa. Antimicrob Agents Chemother. 1990;34:52-57.
Escherichia coli RTEM \g=b\-lactamase by clavulanic acid. Biochemistry. 77. Jordan DC, Mallory HDC. Site of action of vancomycin on Staphylococ-
1978;17:2180-2184. cus aureus. Antimicrob Agents Chemother. 1964;1963:489-494.
48. Fisher J, Charnas RL, Bradley SM, et al. Inactivation of the RTEM \g=b\ x=req-\ 78. Watanakunakorn C. The antibacterial action of vancomycin. Rev Infect
lactamase from Escherichia coli: interaction of penam sulfones with enzyme. Dis. 1981;3:S210-S215.
Biochemistry. 1981;20:2726-2731. 79. Geraci JE, Hermans PE. Vancomycin. Mayo Clin Proc. 1983;58:88-91.
49. Parker RH, Eggleston M. \g=b\-Lactamaseinhibitors: another approach to 80. Leclercq R, Derlot E, Duval J, et al. Plasmid-mediated resistance to
overcoming antimicrobial resistance. Infect Control. 1987;8:36-40. vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med.
50. Moosdeen F, Keeble J, Williams JD. Induction/inhibition of chromosom- 1988;319:157-161.
al \g=b\-lactamaseby \g=b\-lactamaseinhibitors. Rev Infect Dis. 1986;8(suppl 5):562\x=req-\ 81. Williamson R, Al-Obeid S, Shlaes JH, et al. Inducible resistance to
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
vancomycin in Enterococcus faecium D366. J Infect Dis. 1989;159:1095-1104. tance in Staphylococcus epidermidis. J Bacteriol. 1970;104:1095-1105.
82. Leclercq R, Derlot E, Weber M, et al. Transferable vancomycin and 115. Burns JL, Mendelman PM, Levy J, et al. A permeability barrier as a
teicoplanin resistance in Enterococcus faecium. Antimicrob Agents Che- mechanism of chloramphenicol resistance in Haemophilus influenzae. Antimi-
mother. 1989;33:10-15. crob Agents Chemother. 1985;27:46-54.
83. Shlaes DM, Bouvet A, Devine C, et al. Inducible, transferable resistance 116. Burns JL, Hedin LA, Lien DM. Chloramphenicol resistance in Pseudo-
to vancomycin in Enterococcus faecalis A256. Antimicrob Agents Chemother. monas cepacia because of decreased permeability. Antimicrob Agents Che-
1989;33:198-203. mother. 1989;33:136-141.
84. Nicas TI, Cole CT, Preston DA, et al. Activity of glycopeptides against 117. Quiros LM, Fidalgo S, Mendez FJ, et al. Novel mechanisms of resis-
vancomycin-resistant gram-positive bacteria. Antimicrob Agents Chemother. tance to lincosamides in Staphylococcus and Arthrobacter spp. Antimicrob
1989;33:1477-1481. Agents Chemother. 1988;32:420-425.
85. Smith JT, Amyes SGB. Bacterial resistance to antifolate chemothera- 118. Weisblum B. Inducible resistance to macrolides, lincosamides, and
peutic agents mediated by plasmids. Br Med Bull. 1984;40:42-46. streptogramin type B antibiotics: the resistance phenotype, its biological diver-
86. Hitchings GH, Burchall JJ. Inhibition of folate biosynthesis and function sity, and structural elements that regulate expression\p=m-\areview. J Antimi-
as a basis for chemotherapy. Adv Enzymol. 1965;27:417-468. crob Chemother. 1985;16(suppl A):63-90.
87. Then RL. Mechanisms of resistance to trimethoprim, the sulfonamides, 119. Courvalin P, Ounissi H, Arthur M. Multiplicity of macrolide-lincosa-
and trimethoprim-sulfamethoxazole. Rev Infect Dis. 1982;4:261-269. mide-streptogramin antibiotic resistance determinants. J Antimicrob Che-
88. Young H-K, Amyes SGB. A new mechanism of plasmid trimethoprim mother. 1985;16(suppl A):91-100.
resistance. J Biol Chem. 1986;261:2503-2505. 120. Cravin GR, Gavin R, Fanning T. The transfer RNA binding site of the
89. Wylie BA, Amyes SGB, Young H-K, et al. Identification of a novel 30 s ribosome and the site of tetracycline inhibition. Symp Quant Biol.
plasmid-encoded dihydrofolate reductase mediating high-level resistance to 1969;34:129-137.
trimethoprim. J Antimicrob Chemother. 1988;22:429-435. 121. Speer BS, Salyers AA. Characterization of a novel tetracycline resis-
90. Burns JL, Lien DM, Hedin LA. Isolation and characterization of dihy- tance that functions only in aerobically grown Escherichia coli. J Bacteriol.
drofolate reductase from trimethoprim-susceptible and trimethoprim-resis- 1988;170:1423-1429.
tant Pseudomonas cepacia. Antimicrob Agents Chemother. 1989;33:1247\x=req-\ 122. Park BH, Levy SB. The cryptic tetracycline resistance determinant on
1251. Tn4400 mediates tetracycline degradation as well as tetracycline efflux. Anti-
91. Aymes SGB, Towner KJ, Carter GI, et al. The type VII dihydrofolate microb Agents Chemother. 1988;32:1797-1800.
reductase: a novel plasmid-encoded trimethoprim-resistant enzyme from 123. Speer BS, Salyers AA. Novel aerobic tetracycline resistance gene that
gram-negative bacteria isolated in Britain. J Antimicrob Chemother. chemically modifies tetracycline. J Bacteriol. 1989;171:148-153.
1989;24:111-119. 124. McMurry L, Petrucci RE, Levy SB. Active efflux of tetracycline
92. Flensburg J, Skold O. Massive overproduction of dihydrofolate reduc- encoded by four genetically different tetracycline resistance determinants in
tase in bacteria as a response to the use of trimethoprim. Eur J Biochem. Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:3974-3977.
1987;162:473-476. 125. Speer BS, Salyers AA. A tetracycline efflux gene on Bacteroides
93. Burchall JJ, Elwell LP, Fling ME. Molecular mechanisms of resistance transposon Tn4400 does not contribute to tetracycline resistance. J Bacteriol.
to trimethoprim. Rev Infect Dis. 1982;4:246-254. 1990;172:292-298.
94. Skold O. R-factor-mediated resistance to sulfonamides by a plasmid- 126. Wolfson JS, Hooper DC. The fluoroquinolones: structures, mechanisms
borne, drug-resistant dihydropteroate synthase. Antimicrob Agents Che- of action and resistance, and spectra of activity in vitro. Antimicrob Agents
mother. 1976;9:49-54. Chemother. 1985;28:581-586.
95. Radstrom P, Swedberg G. RSF1010 and a conjugative plasmid contain 127. Forward KR, Harding GKM, Gray GJ, et al. Comparative activities of
SulII, one of two known genes for plasmid-borne sulfonamide resistance dihy- norfloxacin and fifteen other antipseudomonal agents against gentamicin-sus-
dropteroate synthase. Antimicrob Agents Chemother. 1988;32:1684-1692. ceptible and -resistant Pseudomonas aeruginosa strains. Antimicrob Agents
96. Davis BD. Mechanism of bactericidal action of aminoglycosides. Micro- Chemother. 1983;24:602-604.
biol Rev. 1987;51:341-350. 128. Wolfson JS, Hooper DC. Norfloxacin: a new targeted fluoroquinolone
97. Phillips I, Shannon K. Aminoglycoside resistance. Br Med Bull. antimicrobial agent. Ann Intern Med. 1988;108:238-251.
1984;40:28-35. 129. Parry MF, Panzer KB, Yukna ME. Quinolone resistance. Am J Med.
98. Ahmad MH, Rechenmacher A, Bock A. Interaction between aminogly- 1989;87(suppl 5A):12-16.
coside uptake and ribosomal resistance mutations. Antimicrob Agents Che- 130. Walker RC, Wright AJ. The quinolones. Mayo Clin Proc. 1987;62:1007\x=req-\
mother. 1980;18:798-806. 1012.
99. Courvalin P, Davies J. Plasmid-mediated aminoglycoside phosphotrans- 131. Smith JT. The mode of action of 4-quinolones and possible mechanisms
ferase of broad substrate range that phosphorylates amikacin. Antimicrob ofresistance. J Antimicrob Chemother. 1986;18(suppl D):21-29.
Agents Chemother. 1977;11:619-624. 132. Hooper DC, Wolfson JS, Souza KS, et al. Genetic and biochemical
100. Lovering AM, White LO, Reeves DS. Identification of individual ami- characterization of norfloxacin resistance in Escherichia coli. Antimicrob
noglycoside-inactivating enzymes in a mixture by HPLC determination of Agents Chemother. 1986;29:639-644.
reaction products. J Antimicrob Chemother. 1986;18:139-144. 133. Hooper DC, Wolfson JS, Ng EY, et al. Mechanisms of action of and
101. Davies J, Smith DI. Plasmid-determined resistance to antimicrobial resistance to ciprofloxacin. Am J Med. 1987;82(suppl 4A):12-20.
agents. Ann Rev Microbiol. 1978;32:469-518. 134. Hrebenda J, Heleszko H, Brzostek K, et al. Mutation affecting resis-
102. Rivera MJ, Castillo J, Martin C, et al. Aminoglycoside-phosphotrans- tance of Escherichia coli K12 to nalidixic acid. J Gen Microbiol. 1985;131:2285\x=req-\
ferases APH(3')-IV and APH(3") synthesized by a strain of Campylobacter 2292.
coli. J Antimicrob Chemother. 1986;18:153-158. 135. Hirai K, Aoyama H, Suzue S, et al. Isolation and characterization of
103. Watanakunakorn C. The prevalence of high-level aminoglycoside resis- norfloxacin-resistant mutants of Escherichia coli K-12. Antimicrob Agents
tance among enterococci isolated from blood cultures during 1980-1988. J Chemother. 1986;30:248-253.
Antimicrob Chemother. 1989;24:63-68. 136. Aoyama H, Sato K, Kato T, et al. Norfloxacin resistance in a clinical
104. Wilson WR, Geraci JE. Treatment of streptococcal infective endocardi- isolate of Escherichia coli. Antimicrob Agents Chemother. 1987;31:1640-1641.
tis. Am J Med. 1985;78(suppl 6B):128-137. 137. Inoue S, Ohue T, Yamagishi J, et al. Mode of incomplete cross-resis-
105. Watanakunakorn C, Bakie C. Synergism of vancomycin-gentamicin tance among pipemidic, piromidic, and nalidixic acids. Antimicrob Agents
and vancomycin-streptomycin against enterococci. Antimicrob Agents Che- Chemother. 1978;14:240-245.
mother. 1973;4:120-124. 138. Sato K, Inoue Y, Fujii T, et al. Purification and properties of DNA
106. Zervos MJ, Terpenning MS, Schaberg DR, et al. High-level aminogly- gyrase from a fluoroquinolone-resistant strain of Escherichia coli. Antimicrob
coside-resistant enterococci. Arch Intern Med. 1987;147:1591-1594. Agents Chemother. 1986;30:777-780.
107. Zervos MJ, Kauffman CA, Therasse PM, et al. Nosocomial infection by 139. Hirai K, Aoyama H, Irikura T, et al. Differences in susceptibility to
gentamicin-resistant Streptococcus faecalis. Ann Intern Med. 1987;106:687\x=req-\ quinolones of outer membrane mutants of Salmonella typhimurium and Es-
691. cherichia coli. Antimicrob Agents Chemother. 1986;29:535-538.
108. Weems JJ, Lowrance JH, Baddour LM, et al. Molecular epidemiology 140. Hooper DC, Wolfson JS, Souza KS, et al. Mechanisms of quinolone
of nosocomial, multiply aminoglycoside resistant Enterococcus faecalis. J An- resistance in Escherichia coli: characterization of nfxB and cfxB, two mutant
timicrob Chemother. 1989;24:121-130. resistance loci decreasing norfloxacin accumulation. Antimicrob Agents Che-
109. Weisberger AS. Inhibition of protein synthesis by chloramphenicol. mother. 1989;33:283-290.
Ann Rev Med. 1967;18:483-494. 141. Hirai K, Suzue S, Irikura T, et al. Mutations producing resistance to
110. Shaw WV. Bacterial resistance to chloramphenicol. Br Med Bull. norfloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother.
1984;40:36-41. 1987;31:582-586.
111. Shaw WV. Chloramphenicol acetyltransferase: enzymology and molec- 142. Legakis NJ, Tzouvelekis LS, Makris A, et al. Outer membrane alter-
ular biology. CRC Crit Rev Biochem. 1983;14:1-46. ations in multiresistant mutants of Pseudomonas aeruginosa. Antimicrob
112. Smith MD, Kelsey MC. Demonstration of a functional variant of chlor- Agents Chemother. 1989;33:124-127.
amphenicol acetyltransferase in Haemophilus influenzae. J Med Microbiol. 143. Maple P, Hamilton-Miller J, Brumfitt W. Ciprofloxacin resistance in
1989;29:263-268. methicillin- and gentamicin-resistant Staphylococcus aureus. Eur J Clin Mi-
113. Leslie AG, Moody PC, Shaw W. Structure of chloramphenicol acetyl- crobiol Infect Dis. 1989;8:622-624.
transferase at 1.75-Aresolution. ProcNatlAcad Sci USA. 1988;85:4133-4137. 144. Schaefler S. Methicillin-resistant strains of Staphylococcus aureus
114. Shaw WV, Bentley DW, Sands L. Mechanism of chloramphenicol resis- resistant to quinolones. J Clin Microbiol. 1989;27:335-336.
Downloaded From: by a University of Toledo Libraries User on 09/28/2018
You might also like
- EOCQ - Ans - 3 BiologyDocument4 pagesEOCQ - Ans - 3 BiologySabQilah25% (12)
- Poisonous Plants and Related Toxins PDFDocument599 pagesPoisonous Plants and Related Toxins PDFJesus Ignacio Briceño PrietoNo ratings yet
- Antibiotic ResistanceDocument32 pagesAntibiotic ResistanceEmine Alaaddinoglu100% (2)
- Beta-lactam antibiotics: Mechanisms of action, resistance, and adverse effects explainedDocument7 pagesBeta-lactam antibiotics: Mechanisms of action, resistance, and adverse effects explainedmarinos zachiotisNo ratings yet
- Resistencia Parte 2Document3 pagesResistencia Parte 2monica perez garciaNo ratings yet
- Microbial PathogenesisDocument9 pagesMicrobial Pathogenesislulipampin012No ratings yet
- Assignment O1Document17 pagesAssignment O1irfanh15951No ratings yet
- Beta-Lactam Antibiotics: Mechanisms of Action and Resistance and Adverse EffectsDocument13 pagesBeta-Lactam Antibiotics: Mechanisms of Action and Resistance and Adverse EffectsellaNo ratings yet
- Mechanisms of Antibiotic Resistance: DR T. Aswani Ndonga MSC Tid I April 2010Document25 pagesMechanisms of Antibiotic Resistance: DR T. Aswani Ndonga MSC Tid I April 2010sushantk862No ratings yet
- Beta 2 NotesDocument14 pagesBeta 2 Notesdita anisya putriNo ratings yet
- MX Resist Gram+Document8 pagesMX Resist Gram+Yorvi hendersson Ilasaca gaonaNo ratings yet
- Antibiotic ClassificationDocument10 pagesAntibiotic ClassificationRipka_uliNo ratings yet
- Mechanisms of Antibiotic Resistance: DR T. Aswani Ndonga MSC Tid I April 2010Document25 pagesMechanisms of Antibiotic Resistance: DR T. Aswani Ndonga MSC Tid I April 2010GAMING MONKEYNo ratings yet
- Antibiotic Resistance PDFDocument6 pagesAntibiotic Resistance PDFGea MarieNo ratings yet
- Antimicrob. Agents Chemother. 2009 Llarrull 4051 63Document13 pagesAntimicrob. Agents Chemother. 2009 Llarrull 4051 63Felipe MNo ratings yet
- Cơ Chế Kháng Thuốc Gram (+) 2Document9 pagesCơ Chế Kháng Thuốc Gram (+) 2Quỳnh Nguyễn Thị NhưNo ratings yet
- Mechanisms of Antimicrobial Resistance in Bacteria PDFDocument8 pagesMechanisms of Antimicrobial Resistance in Bacteria PDFJuan Pablo Ribón GómezNo ratings yet
- Ijerph 20 01900 v2Document11 pagesIjerph 20 01900 v2edisonballaNo ratings yet
- Resistance To Antibacterial DrugsDocument6 pagesResistance To Antibacterial Drugshassan hanicNo ratings yet
- Pharmacology - Antibiotics - Cell Wall & Membrane Inhibit...Document2 pagesPharmacology - Antibiotics - Cell Wall & Membrane Inhibit...Saurabh Singh RajputNo ratings yet
- NBHS1303 Function of BacteriacidalDocument4 pagesNBHS1303 Function of BacteriacidalJejai AnfieldNo ratings yet
- Antibacterial Resistance: DR Ance Roslina.,M.Kes Bagian Mikrobiologi Fk-Umsu 2019Document45 pagesAntibacterial Resistance: DR Ance Roslina.,M.Kes Bagian Mikrobiologi Fk-Umsu 2019RahmiNo ratings yet
- Shakil 2007Document10 pagesShakil 2007Leila RaNo ratings yet
- Antimicrobial ActionDocument26 pagesAntimicrobial ActionAnne CabreraNo ratings yet
- Protein Science - 2020 - Stogios - Molecular Mechanisms of Vancomycin ResistanceDocument16 pagesProtein Science - 2020 - Stogios - Molecular Mechanisms of Vancomycin ResistanceRobert StryjakNo ratings yet
- Staphylococcus: S. Aureus Strains of IntermediateDocument9 pagesStaphylococcus: S. Aureus Strains of IntermediateRoscelie KhoNo ratings yet
- Pared Celular - Bacteriana - Protein Science - 2019 - FisherDocument18 pagesPared Celular - Bacteriana - Protein Science - 2019 - FisherJordi VirgenNo ratings yet
- Cell Wall Inhibitors: Zarqa University Pharmacy School Clinical Pharmacy and Therapeutics Department Pharmacology IIIDocument45 pagesCell Wall Inhibitors: Zarqa University Pharmacy School Clinical Pharmacy and Therapeutics Department Pharmacology IIIAla'a AL AqrabawyNo ratings yet
- Antibiotic Resistance and Extended Spectrum Beta-Lactamases: Types, Epidemiology and TreatmentDocument12 pagesAntibiotic Resistance and Extended Spectrum Beta-Lactamases: Types, Epidemiology and TreatmentyomnayasminNo ratings yet
- Gram-Positive Three-Component Antimicrobial Peptide-Sensing SystemDocument6 pagesGram-Positive Three-Component Antimicrobial Peptide-Sensing SystemLeeya ComotNo ratings yet
- Courvalin 1996Document9 pagesCourvalin 1996Miy AichNo ratings yet
- Phar - Cell Wall Inhibitor - Lec 7 - TheoDocument20 pagesPhar - Cell Wall Inhibitor - Lec 7 - Theoسلام شاكر حميد جميل 6506No ratings yet
- Antimicrobials and VirusesDocument21 pagesAntimicrobials and Virusesclyde clarionNo ratings yet
- Mechanism of Action of Antibiotics: Subject:Pharmaceutical Microbiology DATE:18/09/2019Document12 pagesMechanism of Action of Antibiotics: Subject:Pharmaceutical Microbiology DATE:18/09/2019rubyNo ratings yet
- Mechanisms of Antibiotic ActionDocument15 pagesMechanisms of Antibiotic ActionMarc Imhotep Cray, M.D.No ratings yet
- Introduction and Overview Antibiotics and Chemotherapeutic Agents Micro 260 Chapter 12 NotesDocument8 pagesIntroduction and Overview Antibiotics and Chemotherapeutic Agents Micro 260 Chapter 12 NotesNEHA DIXITNo ratings yet
- Antimicrobial Chemotherapy Part 2Document27 pagesAntimicrobial Chemotherapy Part 2Samara ZahidNo ratings yet
- Idowu 2020Document9 pagesIdowu 2020Sherwan HusseinNo ratings yet
- AntibioticsDocument5 pagesAntibioticsSneeha VeerakumarNo ratings yet
- Antibiotics Classification GuideDocument16 pagesAntibiotics Classification GuideFarida CitraNo ratings yet
- Bacteria and Bacterial Diseases Lecture NotesDocument8 pagesBacteria and Bacterial Diseases Lecture NotesPowell KitagwaNo ratings yet
- Classification and Characteristics of Antibiotics OoQeaTSDocument7 pagesClassification and Characteristics of Antibiotics OoQeaTSvenkybyagari1129No ratings yet
- Staphylococci antigenic structures and virulence factorsDocument26 pagesStaphylococci antigenic structures and virulence factorsTayyaba TahiraNo ratings yet
- 12 2017 Beta-Lactamases A Focus Current ChallengesDocument15 pages12 2017 Beta-Lactamases A Focus Current ChallengesAlisonNo ratings yet
- β-Lactamases and β-Lactamase Inhibitors in the 21st CenturyDocument29 pagesβ-Lactamases and β-Lactamase Inhibitors in the 21st CenturyMarisol Estefany FarfanNo ratings yet
- stapleton-taylor-2002-methicillin-resistance-in-staphylococcus-aureus-mechanisms-and-modulationDocument16 pagesstapleton-taylor-2002-methicillin-resistance-in-staphylococcus-aureus-mechanisms-and-modulationNyomantrianaNo ratings yet
- pdfDocument3 pagespdfAriana Valentina RodriguezNo ratings yet
- Artículo DefensinasDocument12 pagesArtículo Defensinasjenith andradeNo ratings yet
- Beta-lactam antibiotics_ Mechanisms of action and resistance and adverse effects - UpToDateDocument13 pagesBeta-lactam antibiotics_ Mechanisms of action and resistance and adverse effects - UpToDateWelisson BarbosaNo ratings yet
- AMR: Antimicrobial Resistance and MechanismsDocument40 pagesAMR: Antimicrobial Resistance and MechanismsKenny NgowiNo ratings yet
- Antibiotic Resistance Mechanisms in Bacteria: Biochemical and Genetic AspectsDocument11 pagesAntibiotic Resistance Mechanisms in Bacteria: Biochemical and Genetic AspectsD Wisam NajmNo ratings yet
- Abiola Muhammad, ADEOSUN Department of Biochemistry, Lead City University, IbadanDocument37 pagesAbiola Muhammad, ADEOSUN Department of Biochemistry, Lead City University, IbadanKhushi PatelNo ratings yet
- Drug Resistance: College of Ashur University Department of Pharmacy PharmacologyDocument12 pagesDrug Resistance: College of Ashur University Department of Pharmacy Pharmacologyمحمود محمدNo ratings yet
- Khameneh 2019Document28 pagesKhameneh 2019Monyet...No ratings yet
- Tigecyclin Review Journal 2012Document12 pagesTigecyclin Review Journal 2012Retha SaragihNo ratings yet
- 10.1007@978 1 59745 180 28 PDFDocument15 pages10.1007@978 1 59745 180 28 PDFAlma AwaliyahNo ratings yet
- Colistin Resistance in Gram-Negative Bacteria Analysed by Five Phenotypic Assays and Inference of The Underlying Genomic MechanismsDocument12 pagesColistin Resistance in Gram-Negative Bacteria Analysed by Five Phenotypic Assays and Inference of The Underlying Genomic MechanismsBruno LopezNo ratings yet
- Beta-Lactam Antibiotics & Other Inhibitors of Cell WallDocument71 pagesBeta-Lactam Antibiotics & Other Inhibitors of Cell WallAlvin LaurenceNo ratings yet
- Abodakpi 2019 PKPDDocument17 pagesAbodakpi 2019 PKPDCarlos Espinoza CobeñasNo ratings yet
- Understanding Antibiotics Classification and MechanismsDocument41 pagesUnderstanding Antibiotics Classification and MechanismsSyed Ali ShahNo ratings yet
- Cellular and Molecular Mechanisms of Inflammation: Receptors of Inflammatory Cells: Structure—Function RelationshipsFrom EverandCellular and Molecular Mechanisms of Inflammation: Receptors of Inflammatory Cells: Structure—Function RelationshipsCharles G. CochraneNo ratings yet
- Tugas Paraphrase - Annisa YohanesDocument1 pageTugas Paraphrase - Annisa YohanesAnnisa YohanesNo ratings yet
- Prevalence of Antibody To Malignant Catarrhal Fever Virus in Wild and Domestic Ruminants by Competitive-Inhibition ElisaDocument8 pagesPrevalence of Antibody To Malignant Catarrhal Fever Virus in Wild and Domestic Ruminants by Competitive-Inhibition ElisaAnnisa YohanesNo ratings yet
- Fvets 07 00477Document24 pagesFvets 07 00477Annisa YohanesNo ratings yet
- Shampoo Therapy CAVD HandoutDocument2 pagesShampoo Therapy CAVD HandoutAnnisa YohanesNo ratings yet
- Semen Cair EditDocument1 pageSemen Cair EditAnnisa YohanesNo ratings yet
- Animal Nutrition: Original Research ArticleDocument7 pagesAnimal Nutrition: Original Research ArticleAnnisa YohanesNo ratings yet
- Animal Nutrition: Hengxiao Zhai, Hong Liu, Shikui Wang, Jinlong Wu, Anna-Maria KluenterDocument8 pagesAnimal Nutrition: Hengxiao Zhai, Hong Liu, Shikui Wang, Jinlong Wu, Anna-Maria KluenterAnnisa YohanesNo ratings yet
- MDocument10 pagesMAnnisa YohanesNo ratings yet
- Silabus TOEFL Bem KMDocument7 pagesSilabus TOEFL Bem KMAnnisa YohanesNo ratings yet
- Presentation Title: Pitch Deck Tagline Can Extend To Two LinesDocument19 pagesPresentation Title: Pitch Deck Tagline Can Extend To Two LinesAnnisa YohanesNo ratings yet
- Thabet 2017Document7 pagesThabet 2017Annisa YohanesNo ratings yet
- Dapus Lap Akhir-1Document2 pagesDapus Lap Akhir-1Annisa YohanesNo ratings yet
- Presentation Title: Pitch Deck Tagline Can Extend To Two LinesDocument19 pagesPresentation Title: Pitch Deck Tagline Can Extend To Two LinesAnnisa YohanesNo ratings yet
- Pacop Violet Pharmaceutical ChemistryDocument70 pagesPacop Violet Pharmaceutical ChemistryAstherielle GalvezNo ratings yet
- Lehane Histamine Scombroid ReportDocument84 pagesLehane Histamine Scombroid ReportGeraldine Pardo MariluzNo ratings yet
- CC2 Lec PrelimDocument9 pagesCC2 Lec PrelimYo Issei HyodonoNo ratings yet
- Toxicology of Newer Insecticides in Small Animals - Wismer and Means 2012Document13 pagesToxicology of Newer Insecticides in Small Animals - Wismer and Means 2012Camyla NunesNo ratings yet
- Nova BiologyDocument410 pagesNova Biologysupherao150% (2)
- Skin Whitening Cosmetics Feedback and Challenges I PDFDocument24 pagesSkin Whitening Cosmetics Feedback and Challenges I PDFSandeep RautNo ratings yet
- Pharmacology StudentnotesDocument55 pagesPharmacology StudentnotesPasalau Daria VasilicaNo ratings yet
- ITC ExplainedDocument6 pagesITC ExplainedShahil AlamNo ratings yet
- A Review On New Target Insulysine (Ide) & Its Inhibitors As Antidiabetics AgentsDocument12 pagesA Review On New Target Insulysine (Ide) & Its Inhibitors As Antidiabetics AgentsTJPRC PublicationsNo ratings yet
- Holiday Assignment - EssayDocument13 pagesHoliday Assignment - EssayElaine ChoiNo ratings yet
- Cell surface area to volume ratio graphDocument28 pagesCell surface area to volume ratio graphikaNo ratings yet
- Digestion & Absorption ProteinsDocument44 pagesDigestion & Absorption ProteinsPutri Indah NirmalasariNo ratings yet
- Biology WS AnswersDocument17 pagesBiology WS AnswersKazeNo ratings yet
- Chapter 8 Lecture-Energy, Enzymes, and Metabolism-MODIFIED2Document49 pagesChapter 8 Lecture-Energy, Enzymes, and Metabolism-MODIFIED2E'Lasia LarkinNo ratings yet
- Dna LigaseDocument14 pagesDna LigaseAysca Fakhira Amalia100% (1)
- Articulo QBDocument11 pagesArticulo QBAlejandra CortesNo ratings yet
- Lecture 6 - Enzymes As Drug TargetsDocument31 pagesLecture 6 - Enzymes As Drug TargetsAlex LesNo ratings yet
- 9700 BIOLOGY: MARK SCHEME For The October/November 2015 SeriesDocument11 pages9700 BIOLOGY: MARK SCHEME For The October/November 2015 SeriesNerminNo ratings yet
- Pone 0033645Document14 pagesPone 0033645mNo ratings yet
- Problem Set 2Document5 pagesProblem Set 2Fran LeeNo ratings yet
- Taro 2 PDFDocument7 pagesTaro 2 PDFNatasya PermataNo ratings yet
- Regulation of Enzyme ActivityDocument26 pagesRegulation of Enzyme ActivityWeird WorldNo ratings yet
- Mock Exam QP Chemistry HL P3Document20 pagesMock Exam QP Chemistry HL P3jayasimhaNo ratings yet
- Antibiotics Classification GuideDocument16 pagesAntibiotics Classification GuideFarida CitraNo ratings yet
- Spe 87470 MSDocument10 pagesSpe 87470 MSRégis OngolloNo ratings yet
- Inhibitory Effects of Scutellaria Barbata D. Don. and Euonymus Alatus Sieb. On Aromatase Activity of Human Leiomyomal CellsDocument15 pagesInhibitory Effects of Scutellaria Barbata D. Don. and Euonymus Alatus Sieb. On Aromatase Activity of Human Leiomyomal CellsantaeuslabsNo ratings yet
- An Overview of Green Corrosion Inhibitors For Sustainable and Environment Friendly Industrial DevelopmentDocument19 pagesAn Overview of Green Corrosion Inhibitors For Sustainable and Environment Friendly Industrial DevelopmentAbdinasir Mohamed AdanNo ratings yet