Professional Documents
Culture Documents
Meiosis: Crossover Junction Endonuclease MUS81 Is An
Uploaded by
popularsoda0 ratings0% found this document useful (0 votes)
32 views8 pagesOriginal Title
MUS81
Copyright
© © All Rights Reserved
Available Formats
PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
32 views8 pagesMeiosis: Crossover Junction Endonuclease MUS81 Is An
Uploaded by
popularsodaCopyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
You are on page 1of 8
Not logged in Talk Contributions Create account Log in
Article Talk Read Edit View history Search Wikipedia
MUS81
From Wikipedia, the free encyclopedia
Main page Crossover junction endonuclease MUS81 is an
MUS81
Contents enzyme that in humans is encoded by the MUS81
Current events gene.[5][6][7] Available structures
Random article PDB Ortholog search: PDBe RCSB
About Wikipedia
In mammalian somatic cells, MUS81 and another
structure specific DNA endonuclease, XPF (ERCC4), List of PDB id codes
Contact us
Donate play overlapping and essential roles in completion of 2MC3, 4P0P, 4P0Q, 4P0R, 4P0S
homologous recombination.[8] The significant overlap in
Contribute Identifiers
function between these enzymes is most likely related to
Help Aliases MUS81, SLX3, MUS81 structure-specific
processing joint molecules such as D-loops and nicked
Learn to edit endonuclease subunit
Holliday junctions.[8]
Community portal External OMIM: 606591 MGI: 1918961 HomoloGene: 5725
Recent changes IDs GeneCards: MUS81
Contents [hide]
Upload file Gene location (Human)
1 Meiosis
Tools 2 Interactions
What links here 3 References
Related changes 4 Further reading
Special pages
Permanent link
Page information Meiosis [edit]
Cite this page Chr. Chromosome 11 (human)[1]
Wikidata item
MUS81 is a component of a minor chromosomal
crossover (CO) pathway in the meiosis of budding
Print/export yeast, plants and vertebrates.[9] However, in the
Download as PDF protozoan Tetrahymena thermophila, MUS81 appears to Band 11q13.1 Start 65,857,126 bp[1]
be part of an essential (if not the predominant) CO
Languages End 65,867,653 bp[1]
pathway.[9] The MUS81 pathway also appears to be the
Татарча/tatarça Gene location (Mouse)
predominant CO pathway in the fission yeast
Українська
Edit links Schizosaccharomyces pombe.[9]
The relationship of the CO pathway to the overall
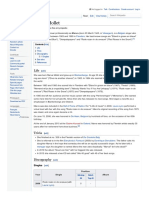
process of meiotic recombination is illustrated in the
accompanying diagram. Recombination during meiosis
is often initiated by a DNA double-strand break (DSB).
During recombination, sections of DNA at the 5' ends of Chr. Chromosome 19 (mouse)[2]
the break are cut away in a process called resection. In
the strand invasion step that follows, an overhanging 3'
end of the broken DNA molecule "invades" the DNA of
Band 19|19 A Start 5,482,345 bp[2]
an homologous chromosome that is not broken forming
End 5,488,402 bp[2]
a displacement loop (D-loop). After strand invasion, the
further sequence of events may follow either of two main RNA expression pattern
pathways, leading to a crossover (CO) or a non-
crossover (NCO) recombinant (see Genetic
recombination). The pathway leading to a CO involves a
double Holliday junction (DHJ) intermediate. Holliday
junctions need to be resolved for CO recombination to
be completed.
MU81-MMS4, in the budding yeast Saccharomyces
cerevisiae, is a DNA structure-selective endonuclease
that cleaves joint DNA molecules formed during More reference expression data
homologous recombination in meiosis and mitosis.[10]
The MUS81-MMS4 endonuclease, although a minor
resolvase for CO formation in S. cerevisiae, is crucial for
limiting chromosome entanglements by suppressing Gene ontology
multiple consecutive recombination events from initiating Molecular • DNA binding
from the same DSB.[11] function • nuclease activity
Mus81 deficient mice have significant meiotic defects • endonuclease activity
• GO:0001948 protein binding
including the failure to repair a subset of DSBs.[12] • 3'-flap endonuclease activity
• metal ion binding
Interactions [edit] • hydrolase activity
• endodeoxyribonuclease activity
MUS81 has been shown to interact with CHEK2.[5] • crossover junction
endodeoxyribonuclease activity
References [edit]
Cellular • nucleoplasm
1. ^a b c GRCh38: Ensembl release 89: component • cell nucleus
ENSG00000172732 - Ensembl, May 2017 • nucleolus
2. ^ a b c GRCm38: Ensembl release 89: • Holliday junction resolvase complex
ENSMUSG00000024906 - Ensembl, May 2017 Biological • DNA recombination
3. ^ "Human PubMed Reference:" . National Center for process • interstrand cross-link repair
Biotechnology Information, U.S. National Library of • DNA catabolic process, endonucleolytic
Medicine. • cellular response to DNA damage
4. ^ "Mouse PubMed Reference:" . National Center for stimulus
Biotechnology Information, U.S. National Library of • response to intra-S DNA damage
Medicine. checkpoint signaling
5. ^ a b Chen XB, Melchionna R, Denis CM, Gaillard PH, • DNA repair
• resolution of meiotic recombination
Blasina A, Van de Weyer I, Boddy MN, Russell P,
intermediates
Vialard J, McGowan CH (Nov 2001). "Human Mus81- • double-strand break repair via break-
associated endonuclease cleaves Holliday junctions in induced replication
vitro". Molecular Cell. 8 (5): 1117–27. • mitotic G2 DNA damage checkpoint
doi:10.1016/S1097-2765(01)00375-6 . • replication fork processing
PMID 11741546 . • intra-S DNA damage checkpoint
6. ^ Constantinou A, Chen XB, McGowan CH, West SC • DNA metabolic process
(Oct 2002). "Holliday junction resolution in human • nucleic acid phosphodiester bond
cells: two junction endonucleases with distinct hydrolysis
substrate specificities" . The EMBO Journal. 21 (20): Sources:Amigo / QuickGO
5577–85. doi:10.1093/emboj/cdf554 .
PMC 129086 . PMID 12374758 .
7. ^ "Entrez Gene: MUS81 MUS81 endonuclease
homolog (S. cerevisiae)" . Orthologs
8. ^ a b Kikuchi K, Narita T, Pham VT, Iijima J, Hirota K, Species Human Mouse
Keka IS, Mohiuddin, Okawa K, Hori T, Fukagawa T, Entrez
Essers J, Kanaar R, Whitby MC, Sugasawa K, 80198 71711
Taniguchi Y, Kitagawa K, Takeda S (2013). "Structure- Ensembl
specific endonucleases xpf and mus81 play ENSG00000172732 ENSMUSG00000024906
overlapping but essential roles in DNA repair by
UniProt
homologous recombination" . Cancer Res. 73 (14): Q96NY9 Q91ZJ0
4362–71. doi:10.1158/0008-5472.CAN-12-3154 .
PMC 3718858 . PMID 23576554 . RefSeq
9. ^ a b c Lukaszewicz A, Howard-Till RA, Loidl J (2013). (mRNA) NM_025128 NM_027877
NM_001350283
"Mus81 nuclease and Sgs1 helicase are essential for
meiotic recombination in a protist lacking a RefSeq
synaptonemal complex" . Nucleic Acids Res. 41 (protein) NP_079404 NP_082153
NP_001337212
(20): 9296–309. doi:10.1093/nar/gkt703 .
PMC 3814389 . PMID 23935123 . Location Chr 11: 65.86 – Chr 19: 5.48 – 5.49 Mb
10. ^ Mukherjee S, Wright WD, Ehmsen KT, Heyer WD (UCSC) 65.87 Mb
(2014). "The Mus81-Mms4 structure-selective PubMed [3] [4]
endonuclease requires nicked DNA junctions to search
undergo conformational changes and bend its DNA Wikidata
substrates for cleavage" . Nucleic Acids Res. 42 View/Edit Human View/Edit Mouse
(10): 6511–22. doi:10.1093/nar/gku265 .
PMC 4041439 . PMID 24744239 .
11. ^ Oke A, Anderson CM, Yam P, Fung JC (2014).
"Controlling meiotic recombinational repair - specifying
the roles of ZMMs, Sgs1 and Mus81/Mms4 in
crossover formation" . PLOS Genet. 10 (10):
e1004690. doi:10.1371/journal.pgen.1004690 .
PMC 4199502 . PMID 25329811 .
PMC 4199502 . PMID 25329811 .
12. ^ Holloway JK, Booth J, Edelmann W, McGowan CH,
Cohen PE (2008). "MUS81 generates a subset of
MLH1-MLH3-independent crossovers in mammalian
meiosis" . PLOS Genet. 4 (9): e1000186.
doi:10.1371/journal.pgen.1000186 . PMC 2525838 .
PMID 18787696 .
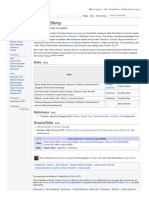
A current model of meiotic recombination, initiated by a double-
strand break or gap, followed by pairing with an homologous
chromosome and strand invasion to initiate the recombinational repair
process. Repair of the gap can lead to crossover (CO) or non-
crossover (NCO) of the flanking regions. CO recombination is thought
to occur by the Double Holliday Junction (DHJ) model, illustrated on the
right, above. NCO recombinants are thought to occur primarily by the
Synthesis Dependent Strand Annealing (SDSA) model, illustrated on
the left, above. Most recombination events appear to be the SDSA type.
Further reading [edit]
Maruyama K, Sugano S (Jan 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic
mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8 . PMID 8125298 .
Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (Oct 1997). "Construction and
characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56.
doi:10.1016/S0378-1119(97)00411-3 . PMID 9373149 .
Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P (Dec 2000). "Damage tolerance protein
Mus81 associates with the FHA1 domain of checkpoint kinase Cds1" . Molecular and Cellular Biology. 20 (23):
8758–66. doi:10.1128/MCB.20.23.8758-8766.2000 . PMC 86503 . PMID 11073977 .
Oğrünç M, Sancar A (Jun 2003). "Identification and characterization of human MUS81-MMS4 structure-specific
endonuclease" . The Journal of Biological Chemistry. 278 (24): 21715–20. doi:10.1074/jbc.M302484200 .
PMID 12686547 .
Ciccia A, Constantinou A, West SC (Jul 2003). "Identification and characterization of the human mus81-eme1
endonuclease" . The Journal of Biological Chemistry. 278 (27): 25172–8. doi:10.1074/jbc.M302882200 .
PMID 12721304 .
Blais V, Gao H, Elwell CA, Boddy MN, Gaillard PH, Russell P, McGowan CH (Feb 2004). "RNA interference inhibition
of Mus81 reduces mitotic recombination in human cells" . Molecular Biology of the Cell. 15 (2): 552–62.
doi:10.1091/mbc.E03-08-0580 . PMC 329235 . PMID 14617801 .
Gao H, Chen XB, McGowan CH (Dec 2003). "Mus81 endonuclease localizes to nucleoli and to regions of DNA
damage in human S-phase cells" . Molecular Biology of the Cell. 14 (12): 4826–34. doi:10.1091/mbc.E03-05-0276 .
PMC 284787 . PMID 14638871 .
Zhang R, Sengupta S, Yang Q, Linke SP, Yanaihara N, Bradsher J, Blais V, McGowan CH, Harris CC (Apr 2005).
"BLM helicase facilitates Mus81 endonuclease activity in human cells" . Cancer Research. 65 (7): 2526–31.
doi:10.1158/0008-5472.CAN-04-2421 . PMID 15805243 .
Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-
Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL,
Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute
J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct
2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8.
doi:10.1038/nature04209 . PMID 16189514 . S2CID 4427026 .
Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K,
Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T,
Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai
K, Isogai T, Sugano S (Jan 2006). "Diversification of transcriptional modulation: large-scale identification and
characterization of putative alternative promoters of human genes" . Genome Research. 16 (1): 55–65.
doi:10.1101/gr.4039406 . PMC 1356129 . PMID 16344560 .
Hiyama T, Katsura M, Yoshihara T, Ishida M, Kinomura A, Tonda T, Asahara T, Miyagawa K (2006).
"Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and
promotes rereplication in human cells" . Nucleic Acids Research. 34 (3): 880–92. doi:10.1093/nar/gkj495 .
PMC 1360746 . PMID 16456034 .
Ii M, Ii T, Brill SJ (Dec 2007). "Mus81 functions in the quality control of replication forks at the rDNA and is involved in
the maintenance of rDNA repeat number in Saccharomyces cerevisiae" . Mutation Research. 625 (1–2): 1–19.
doi:10.1016/j.mrfmmm.2007.04.007 . PMC 2100401 . PMID 17555773 .
Nomura Y, Adachi N, Koyama H (Oct 2007). "Human Mus81 and FANCB independently contribute to repair of DNA
damage during replication". Genes to Cells. 12 (10): 1111–22. doi:10.1111/j.1365-2443.2007.01124.x .
PMID 17903171 . S2CID 33382415 .
Categories: Genes on human chromosome 11
This page was last edited on 11 October 2020, at 11:32 (UTC).
Text is available under the Creative Commons Attribution-ShareAlike License; additional terms may apply. By using this site, you
agree to the Terms of Use and Privacy Policy. Wikipedia® is a registered trademark of the Wikimedia Foundation, Inc., a non-profit
organization.
Privacy policy About Wikipedia Disclaimers Contact Wikipedia Mobile view Developers Statistics Cookie statement
You might also like
- AbstractDocument35 pagesAbstractITZELNo ratings yet
- Cold Spring Harb Perspect Biol-2015-Ohkura-a015859Document15 pagesCold Spring Harb Perspect Biol-2015-Ohkura-a015859wafiqahNo ratings yet
- Acb 43 97Document13 pagesAcb 43 97maryjoseNo ratings yet
- Mamta Nehra, Et AlDocument15 pagesMamta Nehra, Et AlTooba ZahraNo ratings yet
- NUO011Document38 pagesNUO011Hello HiNo ratings yet
- (Reviews of Reproduction) Mitochondrial DNA in Mammalian ReproductionDocument11 pages(Reviews of Reproduction) Mitochondrial DNA in Mammalian ReproductionLes BennettNo ratings yet
- Gky 456Document28 pagesGky 456Mauro Porcel de PeraltaNo ratings yet
- Minireview: The Nature of Meiotic Chromosome Dynamics and Recombination in Budding YeastDocument2 pagesMinireview: The Nature of Meiotic Chromosome Dynamics and Recombination in Budding YeastPrasetyaNo ratings yet
- Review Article: Finding The Correct Partner: The Meiotic CourtshipDocument14 pagesReview Article: Finding The Correct Partner: The Meiotic CourtshipveronicaNo ratings yet
- Molecular Biology PrimerDocument32 pagesMolecular Biology Primermergun919No ratings yet
- TMP 7 AFFDocument4 pagesTMP 7 AFFFrontiersNo ratings yet
- Evolution and Inheritance of Animal Mitochondrial DNA: Rules and ExceptionsDocument7 pagesEvolution and Inheritance of Animal Mitochondrial DNA: Rules and ExceptionsIntan Sartika RiskyNo ratings yet
- Gene Editing, Epigenetic, Cloning and TherapyFrom EverandGene Editing, Epigenetic, Cloning and TherapyRating: 4 out of 5 stars4/5 (1)
- Mitochondrial Genomes Anything Goes Trends Genet 1Document9 pagesMitochondrial Genomes Anything Goes Trends Genet 1Alexandre Guitart LuengoNo ratings yet
- Schenkel Et Al 2023 A Dedicated Cytoplasmic Container Collects Extrachromosomal Dna Away From The Mammalian NucleusDocument21 pagesSchenkel Et Al 2023 A Dedicated Cytoplasmic Container Collects Extrachromosomal Dna Away From The Mammalian NucleusGuilhermeNo ratings yet
- Clase 6Document58 pagesClase 6MATEO SEBASTIAN RODRIGUEZ TORRESNo ratings yet
- La Souris, La Mouche Et L'Homme A. Human Genome Project: B. DNA Computers/DNA Nanorobots: C. PhylogenomicsDocument12 pagesLa Souris, La Mouche Et L'Homme A. Human Genome Project: B. DNA Computers/DNA Nanorobots: C. PhylogenomicsFadhili DungaNo ratings yet
- 1 s2.0 S0070215322000886 MainDocument36 pages1 s2.0 S0070215322000886 Mainm1981pomNo ratings yet
- Mutation Chromosome Change in Number and StructureDocument53 pagesMutation Chromosome Change in Number and StructureyukbhuviNo ratings yet
- Lecture 2016 - Cell NucleusDocument17 pagesLecture 2016 - Cell NucleusKhalil RehmanNo ratings yet
- Lec 12Document4 pagesLec 12Jastine Ella Maxidel SimporiosNo ratings yet
- The Role of Nucleic Acids in HeredityDocument4 pagesThe Role of Nucleic Acids in HeredityRaluca GladcencoNo ratings yet
- Studying Phase Separation in Confinement: SciencedirectDocument12 pagesStudying Phase Separation in Confinement: SciencedirectJuliana VilachãNo ratings yet
- CellDocument10 pagesCelljuju on the beatNo ratings yet
- Recombinanat DNA TechnologyDocument50 pagesRecombinanat DNA Technologymulatu mokononNo ratings yet
- Deinum 2016Document14 pagesDeinum 2016Tahiri AbdelilahNo ratings yet
- Mechanism and Regulation of Human Non-Homologous Dna End-JoiningDocument9 pagesMechanism and Regulation of Human Non-Homologous Dna End-JoiningAbi Wombat MendiolitaNo ratings yet
- Cells 08 00912 v2Document19 pagesCells 08 00912 v2dffNo ratings yet
- Mitochondria More Than JustDocument10 pagesMitochondria More Than JustCarla MONo ratings yet
- Unit 2. Molecular GeneticsDocument53 pagesUnit 2. Molecular GeneticsRuben Ruiz MartinNo ratings yet
- Reviews: The Cell Biology of Mitochondrial Membrane DynamicsDocument21 pagesReviews: The Cell Biology of Mitochondrial Membrane DynamicsSaidahNo ratings yet
- Lane Bioessays Mitonuclear MatchDocument10 pagesLane Bioessays Mitonuclear Match22194No ratings yet
- BTB 103 Lecture 32Document10 pagesBTB 103 Lecture 32Anusha OfficialNo ratings yet
- Note 1492433946Document23 pagesNote 1492433946Priyanshi choudharyNo ratings yet
- 03 Sewage Treatment With Anammox (KARTAL)Document2 pages03 Sewage Treatment With Anammox (KARTAL)Eduardo FleckNo ratings yet
- Mitochondria Impaired Mitochondrial Translation in Human DiseaseDocument8 pagesMitochondria Impaired Mitochondrial Translation in Human DiseaseParvinder McCartneyNo ratings yet
- Cold Spring Harb Perspect Biol-2015-Lam-a016634Document26 pagesCold Spring Harb Perspect Biol-2015-Lam-a016634Turtle ArtNo ratings yet
- The Genome Loading Model For The Origin and MainteDocument13 pagesThe Genome Loading Model For The Origin and MainteShopnil SarkarNo ratings yet
- Article: Cell-Cycle Kinases Coordinate The Resolution of Recombination Intermediates With Chromosome SegregationDocument16 pagesArticle: Cell-Cycle Kinases Coordinate The Resolution of Recombination Intermediates With Chromosome SegregationManishGautamNo ratings yet
- Topical Guidebook For GCE O Level Biology 3 Part 2From EverandTopical Guidebook For GCE O Level Biology 3 Part 2Rating: 5 out of 5 stars5/5 (1)
- Full Download Genetics A Conceptual Approach 6th Edition Pierce Solutions ManualDocument35 pagesFull Download Genetics A Conceptual Approach 6th Edition Pierce Solutions Manualdopemorpheanwlzyv100% (38)
- Beyond Tethering and The Lem Domain: Mscellaneous Functions of The Inner Nuclear Membrane Lem2Document10 pagesBeyond Tethering and The Lem Domain: Mscellaneous Functions of The Inner Nuclear Membrane Lem2Lê Khánh ToànNo ratings yet
- Module 2. Molecular Biology (Week 4-7)Document15 pagesModule 2. Molecular Biology (Week 4-7)Nanette MangloNo ratings yet
- Giasanti and Scovassi, 2008Document8 pagesGiasanti and Scovassi, 2008Khate Catherine AsuncionNo ratings yet
- Pertumbuhan SelDocument37 pagesPertumbuhan SelbeniNo ratings yet
- Molecular Genetics Chapter 8Document49 pagesMolecular Genetics Chapter 8byunus88No ratings yet
- SM Research Paper-2Document17 pagesSM Research Paper-2sourobh MAjiNo ratings yet
- 10 1002@anie 202011711Document10 pages10 1002@anie 202011711Abhijeet MohantyNo ratings yet
- HOMOLOGOUS AND NON HOMOLOGOUS RECOMBINATION by Preeti Saini 21mslsmm06Document7 pagesHOMOLOGOUS AND NON HOMOLOGOUS RECOMBINATION by Preeti Saini 21mslsmm06Preeti SainiNo ratings yet
- Epigenetics - It's Not Just Genes That Make Us - British Society For Cell BiologDocument12 pagesEpigenetics - It's Not Just Genes That Make Us - British Society For Cell BiologRidhima AroraNo ratings yet
- 2009 SolutionsDocument21 pages2009 SolutionsSonal Power UnlimitdNo ratings yet
- New Syllabus AnswerDocument24 pagesNew Syllabus AnswerKhaled DabourNo ratings yet
- Addis Ababa University Institute of Biotechnology Presentation Assignment For The Course Advanced Molecular Biology (BIOT 801)Document23 pagesAddis Ababa University Institute of Biotechnology Presentation Assignment For The Course Advanced Molecular Biology (BIOT 801)HelenNo ratings yet
- Dna Replication and MutationDocument4 pagesDna Replication and MutationmichsantosNo ratings yet
- HHS Public Access: Eukaryotic DNA Replication ForkDocument26 pagesHHS Public Access: Eukaryotic DNA Replication ForkGustavo MoraNo ratings yet
- Cell DivisionDocument28 pagesCell DivisionPalagiri MadhuNo ratings yet
- 2012 - MalhotraDocument19 pages2012 - MalhotraCarlNo ratings yet
- Biological Data Paper 1Document6 pagesBiological Data Paper 1Medha PatelNo ratings yet
- A Histone CycleDocument26 pagesA Histone CycleliaNo ratings yet
- Beata WickhamiDocument1 pageBeata WickhamipopularsodaNo ratings yet
- Whipple-Angell-Bennett House: See AlsoDocument2 pagesWhipple-Angell-Bennett House: See AlsopopularsodaNo ratings yet
- 1734 in Science PDFDocument2 pages1734 in Science PDFpopularsodaNo ratings yet
- Austromuellera: ReferencesDocument18 pagesAustromuellera: ReferencespopularsodaNo ratings yet
- Aluminium-Air BatteryDocument6 pagesAluminium-Air BatterypopularsodaNo ratings yet
- UMBC Retrievers Men's LacrosseDocument4 pagesUMBC Retrievers Men's LacrossepopularsodaNo ratings yet
- Yalta Municipality: Administrative and Municipal StatusDocument3 pagesYalta Municipality: Administrative and Municipal StatuspopularsodaNo ratings yet
- The Edge of The AbyssDocument1 pageThe Edge of The AbysspopularsodaNo ratings yet
- Trade WindsDocument9 pagesTrade WindspopularsodaNo ratings yet
- Re-Animator (Album) : Recording and ProductionDocument5 pagesRe-Animator (Album) : Recording and ProductionpopularsodaNo ratings yet
- Tere MarichalDocument2 pagesTere MarichalpopularsodaNo ratings yet
- Stochastic Petri NetDocument2 pagesStochastic Petri NetpopularsodaNo ratings yet
- Sin-Eater: AttestationsDocument9 pagesSin-Eater: AttestationspopularsodaNo ratings yet
- Sonya's StoryDocument2 pagesSonya's StorypopularsodaNo ratings yet
- Tarachodes NatalensisDocument1 pageTarachodes NatalensispopularsodaNo ratings yet
- Kulkumish, CaliforniaDocument1 pageKulkumish, CaliforniapopularsodaNo ratings yet
- Pa'amei TashazDocument1 pagePa'amei TashazpopularsodaNo ratings yet
- Milan Milutinovic (Footballer)Document1 pageMilan Milutinovic (Footballer)popularsodaNo ratings yet
- Raid On KhatabaDocument6 pagesRaid On KhatabapopularsodaNo ratings yet
- PlatyspiraDocument1 pagePlatyspirapopularsodaNo ratings yet
- Mourad Boudjellal: 2006-2020: President of The Club (TRUC)Document2 pagesMourad Boudjellal: 2006-2020: President of The Club (TRUC)popularsodaNo ratings yet
- Mike Craig (Field Hockey)Document1 pageMike Craig (Field Hockey)popularsodaNo ratings yet
- Marva Mollet: Marva Mollet, Known Professionally As Marva (Born 23 March 1943, inDocument2 pagesMarva Mollet: Marva Mollet, Known Professionally As Marva (Born 23 March 1943, inpopularsodaNo ratings yet
- Felix HamrinDocument2 pagesFelix HamrinpopularsodaNo ratings yet
- Kenneth S. WarrenDocument1 pageKenneth S. WarrenpopularsodaNo ratings yet
- Francisco de La Cerda CórdobaDocument2 pagesFrancisco de La Cerda CórdobapopularsodaNo ratings yet
- J. Parnell ThomasDocument3 pagesJ. Parnell ThomaspopularsodaNo ratings yet
- Eutreta FenestrataDocument1 pageEutreta FenestratapopularsodaNo ratings yet
- Glenn Kinsey: Children's ITVDocument1 pageGlenn Kinsey: Children's ITVpopularsodaNo ratings yet
- Aizen Myo Love Passion & Lust Reiki - GabrielaDocument19 pagesAizen Myo Love Passion & Lust Reiki - GabrielaNadezdaNo ratings yet
- Nonlinear Dynamics in EquilibriumDocument11 pagesNonlinear Dynamics in EquilibriumCosmo ChouNo ratings yet
- Takagi and TogakureDocument1 pageTakagi and TogakureSean AskewNo ratings yet
- Credit To Ariatranslation at Lollipop! Lyrics Credit Tracks 4, 8, & 10 To After - 12 - Am at Lollipop! LyricsDocument22 pagesCredit To Ariatranslation at Lollipop! Lyrics Credit Tracks 4, 8, & 10 To After - 12 - Am at Lollipop! Lyricsisabella_sakuraiNo ratings yet
- 02 AinuDocument8 pages02 AinuGus DurNo ratings yet
- Lista de Empresas ISO 27001Document8 pagesLista de Empresas ISO 27001magnodiasfarahNo ratings yet
- Body Language IIDocument4 pagesBody Language IIFmga Ga0% (1)
- Japanese Nouns: Japanese English Number GenderDocument2 pagesJapanese Nouns: Japanese English Number GenderSteven Bong SabarezaNo ratings yet
- White Belt TerminologyDocument1 pageWhite Belt Terminologyramkannan18No ratings yet
- Hiragana - ひらがな: CombinationsDocument10 pagesHiragana - ひらがな: CombinationsLuis Carlos SalazarNo ratings yet
- Japan Karate Do HakuaDocument76 pagesJapan Karate Do Hakuabrendan lanzaNo ratings yet
- Dark Souls and The Hero's JourneyDocument13 pagesDark Souls and The Hero's JourneyAndrew J. QuesenberryNo ratings yet
- Combat Arts Archive Vintage Martial Arts Book Catalogue Summer 2009 No. 57Document24 pagesCombat Arts Archive Vintage Martial Arts Book Catalogue Summer 2009 No. 57joebirdNo ratings yet
- A Fresh Look at State ShintoDocument16 pagesA Fresh Look at State ShintoSven UstintsevNo ratings yet
- Yashi 2017Document2 pagesYashi 2017api-362876206No ratings yet
- The Five Star Stories Movie.Document17 pagesThe Five Star Stories Movie.xzaydenNo ratings yet
- Lesson 11 Fola 001Document9 pagesLesson 11 Fola 001Arvilyn ArabellaNo ratings yet
- JapDocument45 pagesJapwhateverNo ratings yet
- Japanese 101Document190 pagesJapanese 101Izza Mae Mejillano AnielNo ratings yet
- Sengoku RPG - Revised Ed PDFDocument337 pagesSengoku RPG - Revised Ed PDFmarco.meduri100% (3)
- Kendo Equipment Manual: (Please Don't Illegally Copy This Book)Document51 pagesKendo Equipment Manual: (Please Don't Illegally Copy This Book)Dragan MitrovicNo ratings yet
- Wchapter 12-Japan - Land of The Rising SunDocument5 pagesWchapter 12-Japan - Land of The Rising Sunapi-29949541233% (3)
- Grade 1 Study Course 2015Document31 pagesGrade 1 Study Course 2015Manish GandhiNo ratings yet
- Daido Eng PDFDocument8 pagesDaido Eng PDFreyesfrNo ratings yet
- Dazai OsamuDocument8 pagesDazai OsamuzamatinNo ratings yet
- MokurokuDocument10 pagesMokurokuAjax StingrayNo ratings yet
- Iwashita Et Al-2017-Journal of Hepato-Biliary-Pancreatic SciencesDocument31 pagesIwashita Et Al-2017-Journal of Hepato-Biliary-Pancreatic SciencesCARLOS ALBERTO PALOMINO FIGUEREDONo ratings yet
- Rokugan Vassal FamiliesDocument21 pagesRokugan Vassal FamiliesGuittonNo ratings yet
- Biodata Professor Hiroshi Ochi: Research FieldDocument4 pagesBiodata Professor Hiroshi Ochi: Research FieldabdirahmatNo ratings yet
- Wanderlei Silva Mauricio Shogun' Rua Antonio Rogerio Nogueira Minotouro'Document26 pagesWanderlei Silva Mauricio Shogun' Rua Antonio Rogerio Nogueira Minotouro'api-3828341100% (1)