Professional Documents
Culture Documents
Gram-Negative Bacteria: Characteristics
Uploaded by
Syimah UmarOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Gram-Negative Bacteria: Characteristics
Uploaded by
Syimah UmarCopyright:
Available Formats
Not logged in Talk Contributions Create account Log in
Article Talk Read Edit View history Search Wikipedia
There is still a lot of local
Malaysian cuisines that
does not have its picture
in Wikipedia:
Join WikiSelera 2021 to
Main page share your local food and
Contents win prizes!
Current events
Random article
About Wikipedia Gram-negative bacteria
Contact us
From Wikipedia, the free encyclopedia
Donate
Contribute Gram-negative
Help bacteria are
Learn to edit bacteria that do
Community portal not retain the
Recent changes crystal violet
Upload file stain used in the
Tools gram-staining
method of
What links here
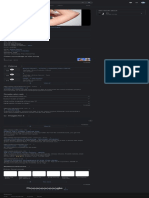
Related changes bacterial Microscopic image of gram-negative
Special pages differentiation.[1] Pseudomonas aeruginosa bacteria
(pink-red rods)
Permanent link They are
Page information characterized by
Cite this page their cell envelopes, which are composed of a thin
Wikidata item
peptidoglycan cell wall sandwiched between an inner
Print/export cytoplasmic cell membrane and a bacterial outer
Download as PDF membrane.
Printable version Gram-negative bacteria are found everywhere, in virtually
all environments on Earth that support life. The gram-
In other projects
negative bacteria include the model organism Escherichia
Wikimedia Commons
coli, as well as many pathogenic bacteria, such as
Languages Pseudomonas aeruginosa, Chlamydia trachomatis, and
العربية Yersinia pestis. They are an important medical challenge,
Español as their outer membrane protects them from many
िहन्दी antibiotics (including penicillin); detergents that would
Bahasa Indonesia normally damage the peptidoglycans of the (inner) cell
Jawa
membrane; and lysozyme, an antimicrobial enzyme
മലയാളം
produced by animals that forms part of the innate immune
Русский
system. Additionally, the outer leaflet of this membrane
!"#
comprises a complex lipopolysaccharide (LPS) whose
lipid A component can cause a toxic reaction when these
38 more bacteria are lysed by immune cells. This toxic reaction
Edit links can include fever, an increased respiratory rate, and low
blood pressure—a life-threatening condition known as
septic shock.[2]
Several classes of antibiotics have been designed to
target gram-negative bacteria, including aminopenicillins,
ureidopenicillins, cephalosporins, beta-lactam-
betalactamase inhibitor combinations (e.g. piperacillin-
tazobactam), Folate antagonists, quinolones, and
carbapenems. Many of these antibiotics also cover gram-
positive organisms. The drugs that specifically target
gram negative organisms include aminoglycosides,
monobactams (aztreonam) and ciprofloxacin.
Contents [hide]
1 Characteristics
2 Classification
3 Taxonomy
3.1 Example species
4 Bacterial transformation
5 Medical treatment
6 Orthographic note
7 See also
8 References
8.1 Notes
9 External links
Characteristics [ edit ]
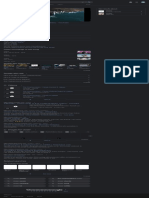
Gram-negative cell wall structure
Gram-negative
bacteria display
Gram-positive and -negative bacteria
are differentiated chiefly by their cell wall
structure
these characteristics:
An inner cell membrane is present (cytoplasmic)
A thin peptidoglycan layer is present (This is much
thicker in gram-positive bacteria)
Has outer membrane containing lipopolysaccharides
(LPS, which consists of lipid A, core polysaccharide,
and O antigen) in its outer leaflet and phospholipids in
the inner leaflet
Porins exist in the outer membrane, which act like
pores for particular molecules
Between the outer membrane and the cytoplasmic
membrane there is a space filled with a concentrated
gel-like substance called periplasm
The S-layer is directly attached to the outer
membrane rather than to the peptidoglycan
If present, flagella have four supporting rings instead
of two
Teichoic acids or lipoteichoic acids are absent
Lipoproteins are attached to the polysaccharide
backbone
Some contain Braun's lipoprotein, which serves as a
link between the outer membrane and the
peptidoglycan chain by a covalent bond
Most, with few exceptions, do not form spores
Classification [ edit ]
Along with cell shape, Gram staining is a rapid diagnostic
tool and once was used to group species at the
subdivision of Bacteria. Historically, the kingdom Monera
was divided into four divisions based on Gram staining:
Firmacutes (+), Gracillicutes (−), Mollicutes (0) and
Mendocutes (var.).[3] Since 1987, the monophyly of the
gram-negative bacteria has been disproven with
molecular studies.[4] However some authors, such as
Cavalier-Smith still treat them as a monophyletic taxon
(though not a clade; his definition of monophyly requires
a single common ancestor but does not require holophyly,
the property that all descendants be encompassed by the
taxon) and refer to the group as a subkingdom
"Negibacteria".[5]
Taxonomy [ edit ]
This section may be too
technical for most readers to
understand. Please help
improve it to make it
understandable to non-experts,
without removing the technical
details. (March 2014) (Learn how
and when to remove this template
message)
Bacteria are traditionally classified based on their Gram-
staining response into the gram-positive and gram-
negative bacteria. Having just one membrane the gram-
positive bacteria are also known as monoderm bacteria,
and gram-negative having two membranes are also
known as diderm bacteria. It was traditionally thought that
the groups represent lineages, i.e. the extra membrane
only evolved once, such that gram-negative bacteria are
more closely related to one another than to any gram-
positive bacteria. While this is often true, the classification
system breaks down in some cases, with lineage
groupings not matching the staining result.[6][7][8][9] Thus,
Gram staining cannot be reliably used to assess familial
relationships of bacteria. Nevertheless, staining often
gives reliable information about the composition of the
cell membrane, distinguishing between the presence or
absence of an outer lipid membrane.[6][10]
Of these two structurally distinct groups of prokaryotic
organisms, monoderm prokaryotes are thought to be
ancestral. Based upon a number of different observations
including that the gram-positive bacteria are the major
reactors to antibiotics and that gram-negative bacteria
are, in general, resistant to them, it has been proposed
that the outer cell membrane in gram-negative bacteria
(diderms) evolved as a protective mechanism against
antibiotic selection pressure.[6][7][10][11] Some bacteria
such as Deinococcus, which stain gram-positive due to
the presence of a thick peptidoglycan layer, but also
possess an outer cell membrane are suggested as
intermediates in the transition between monoderm (gram-
positive) and diderm (gram-negative) bacteria.[6][11] The
diderm bacteria can also be further differentiated between
simple diderms lacking lipopolysaccharide (LPS); the
archetypical diderm bacteria, in which the outer cell
membrane contains lipopolysaccharide; and the diderm
bacteria, in which the outer cell membrane is made up of
mycolic acid (e. g. Mycobacterium).[8][9][11][12]
The conventional LPS-diderm group of gram-negative
bacteria (e.g., Proteobacteria, Aquificae, Chlamydiae,
Bacteroidetes, Chlorobi, Cyanobacteria, Fibrobacteres,
Verrucomicrobia, Planctomycetes, Spirochetes,
Acidobacteria; "Hydrobacteria") are uniquely identified by
a few conserved signature indel (CSI) in the HSP60
(GroEL) protein. In addition, a number of bacterial taxa
(including Negativicutes, Fusobacteria, Synergistetes,
and Elusimicrobia) that are either part of the phylum
Firmicutes (a monoderm group) or branches in its
proximity are also found to possess a diderm cell
structure.[9][11][12] They lack the GroEL signature.[11] The
presence of this CSI in all sequenced species of
conventional lipopolysaccharide-containing gram-
negative bacterial phyla provides evidence that these
phyla of bacteria form a monophyletic clade and that no
loss of the outer membrane from any species from this
group has occurred.[11]
Example species [ edit ]
The proteobacteria are a major phylum of gram-negative
bacteria, including Escherichia coli (E. coli), Salmonella,
Shigella, and other Enterobacteriaceae, Pseudomonas,
Moraxella, Helicobacter, Stenotrophomonas, Bdellovibrio,
acetic acid bacteria, Legionella etc. Other notable groups
of gram-negative bacteria include the cyanobacteria,
spirochaetes, green sulfur, and green non-sulfur bacteria.
Medically relevant gram-negative cocci include the four
types that cause a sexually transmitted disease
(Neisseria gonorrhoeae), a meningitis (Neisseria
meningitidis), and respiratory symptoms (Moraxella
catarrhalis, Haemophilus influenzae).
Medically relevant gram-negative bacilli include a
multitude of species. Some of them cause primarily
respiratory problems (Klebsiella pneumoniae, Legionella
pneumophila, Pseudomonas aeruginosa), primarily
urinary problems (Escherichia coli, Proteus mirabilis,
Enterobacter cloacae, Serratia marcescens), and
primarily gastrointestinal problems (Helicobacter pylori,
Salmonella enteritidis, Salmonella typhi).
Gram-negative bacteria associated with hospital-acquired
infections include Acinetobacter baumannii, which cause
bacteremia, secondary meningitis, and ventilator-
associated pneumonia in hospital intensive-care units.
Bacterial transformation [ edit ]
Transformation is one of three processes for horizontal
gene transfer, in which exogenous genetic material
passes from bacterium to another, the other two being
conjugation (transfer of genetic material between two
bacterial cells in direct contact) and transduction
(injection of foreign DNA by a bacteriophage virus into the
host bacterium).[13] In transformation, the genetic material
passes through the intervening medium, and uptake is
completely dependent on the recipient bacterium.[13]
As of 2014 about 80 species of bacteria were known to
be capable of transformation, about evenly divided
between gram-positive and gram-negative bacteria; the
number might be an overestimate since several of the
reports are supported by single papers.[13]
Transformation has been studied in medically important
gram-negative bacteria species such as Helicobacter
pylori, Legionella pneumophila, Neisseria meningitidis,
Neisseria gonorrhoeae, Haemophilus influenzae and
Vibrio cholerae.[14] It has also been studied in gram-
negative species found in soil such as Pseudomonas
stutzeri, Acinetobacter baylyi, and gram-negative plant
pathogens such as Ralstonia solanacearum and Xylella
fastidiosa.[14]
Medical treatment [ edit ]
This section needs additional
citations for verification.
Please help improve this article
by adding citations to reliable
sources. Unsourced material
may be challenged and
removed. (April 2013) (Learn how
and when to remove this template
message)
One of the several unique characteristics of gram-
negative bacteria is the structure of the bacterial outer
membrane. The outer leaflet of this membrane comprises
a complex lipopolysaccharide (LPS) whose lipid portion
acts as an endotoxin. If gram-negative bacteria enter the
circulatory system, the LPS can cause a toxic reaction.
This results in fever, an increased respiratory rate, and
low blood pressure. This may lead to life-threatening
septic shock.[2]
The outer membrane protects the bacteria from several
antibiotics, dyes, and detergents that would normally
damage either the inner membrane or the cell wall (made
of peptidoglycan). The outer membrane provides these
bacteria with resistance to lysozyme and penicillin. The
periplasmic space (space between the two cell
membranes) also contains enzymes which break down or
modify antibiotics. Drugs commonly used to treat gram
negative infections include amino, carboxy and ureido
penicillins (ampicillin, amoxicillin, pipercillin, ticarcillin)
these drugs may be combined with beta-lactamase
inhibitors to combat the presence of enzymes that can
digest these drugs (known as beta-lactamases) in the
peri-plasmic space. Other classes of drugs that have
gram negative spectrum include cephalosporins,
monobactams (aztreonam), aminogylosides, quinolones,
macrolides, chloramphenicol, folate antagonists, and
carbapenems.[15]
The pathogenic capability of gram-negative bacteria is
often associated with certain components of their
membrane, in particular, the LPS.[1] In humans, the
presence of LPS triggers an innate immune response,
activating the immune system and producing cytokines
(hormonal regulators). Inflammation is a common
reaction to cytokine production, which can also produce
host toxicity. The innate immune response to LPS,
however, is not synonymous with pathogenicity, or the
ability to cause disease.[citation needed]
Orthographic note [ edit ]
The adjectives Gram-positive and Gram-negative derive
from the surname of Hans Christian Gram, a Danish
bacteriologist; as eponymous adjectives, their initial letter
can be either capital G or lower-case g, depending on
which style guide (e.g., that of the CDC), if any, governs
the document being written.[16] This is further explained
at Gram staining § Orthographic note.
See also [ edit ]
Gram-variable and gram-indeterminate bacteria
Outer membrane receptor
References [ edit ]
This article incorporates public domain material from
the NCBI document: "Science Primer" .
Notes [ edit ]
1. ^ a b Baron S, Salton MR, Kim KS (1996).
"Structure" . In Baron S, et al. (eds.). Baron's Medical
Microbiology (4th ed.). Univ of Texas Medical Branch.
ISBN 978-0-9631172-1-2. PMID 21413343 .
2. ^ a b Pellitier LL Jr, "Microbiology of the Circulatory
System" "NCBI Bookshelf", April 18, 2017
3. ^ Gibbons, N. E.; Murray, R. G. E. (1978). "Proposals
Concerning the Higher Taxa of Bacteria" .
International Journal of Systematic Bacteriology. 28
(1): 1–6. doi:10.1099/00207713-28-1-1 .
4. ^ Woese CR (June 1987). "Bacterial evolution" .
Microbiol. Rev. 51 (2): 221–71.
doi:10.1128/MMBR.51.2.221-271.1987 .
PMC 373105 . PMID 2439888 .
5. ^ Cavalier-Smith, T. (2006). "Rooting the tree of life by
transition analyses" . Biol. Direct. 1: 19.
doi:10.1186/1745-6150-1-19 . PMC 1586193 .
PMID 16834776 .
6. ^ a b c d Gupta, RS (December 1998). "Protein
phylogenies and signature sequences: A reappraisal of
evolutionary relationships among archaebacteria,
eubacteria, and eukaryotes" . Microbiol. Mol. Biol.
Rev. 62 (4): 1435–91. doi:10.1128/MMBR.62.4.1435-
1491.1998 . PMC 98952 . PMID 9841678 .
7. ^ a b Gupta RS (2000). "The natural evolutionary
relationships among prokaryotes" (PDF). Crit. Rev.
Microbiol. 26 (2): 111–31.
CiteSeerX 10.1.1.496.1356 .
doi:10.1080/10408410091154219 .
PMID 10890353 .
8. ^ a b Desvaux M, Hébraud M, Talon R, Henderson IR
(April 2009). "Secretion and subcellular localizations of
bacterial proteins: a semantic awareness issue".
Trends Microbiol. 17 (4): 139–45.
doi:10.1016/j.tim.2009.01.004 . PMID 19299134 .
9. ^ a b c Sutcliffe IC (October 2010). "A phylum level
perspective on bacterial cell envelope architecture".
Trends Microbiol. 18 (10): 464–70.
doi:10.1016/j.tim.2010.06.005 . PMID 20637628 .
10. ^ a b Gupta RS (August 1998). "What are
archaebacteria: life's third domain or monoderm
prokaryotes related to gram-positive bacteria? A new
proposal for the classification of prokaryotic
organisms" . Mol. Microbiol. 29 (3): 695–707.
doi:10.1046/j.1365-2958.1998.00978.x .
PMID 9723910 .
11. ^ a b c d e f Gupta RS (August 2011). "Origin of diderm
(gram-negative) bacteria: antibiotic selection pressure
rather than endosymbiosis likely led to the evolution of
bacterial cells with two membranes" . Antonie van
Leeuwenhoek. 100 (2): 171–82. doi:10.1007/s10482-
011-9616-8 . PMC 3133647 . PMID 21717204 .
12. ^ a b Marchandin H, Teyssier C, Campos J, Jean-Pierre
H, Roger F, Gay B, Carlier JP, Jumas-Bilak E (June
2010). "Negativicoccus succinicivorans gen. nov., sp.
nov., isolated from human clinical samples, emended
description of the family Veillonellaceae and
description of Negativicutes classis nov.,
Selenomonadales ord. nov. and Acidaminococcaceae
fam. nov. in the bacterial phylum Firmicutes" . Int. J.
Syst. Evol. Microbiol. 60 (Pt 6): 1271–9.
doi:10.1099/ijs.0.013102-0 . PMID 19667386 .
13. ^ a b c Johnston C, Martin B, Fichant G, Polard P,
Claverys JP (2014). "Bacterial transformation:
distribution, shared mechanisms and divergent
control". Nat. Rev. Microbiol. 12 (3): 181–96.
doi:10.1038/nrmicro3199 . PMID 24509783 .
14. ^ a b Seitz P, Blokesch M (2013). "Cues and regulatory
pathways involved in natural competence and
transformation in pathogenic and environmental Gram-
negative bacteria" . FEMS Microbiol. Rev. 37 (3):
336–63. doi:10.1111/j.1574-6976.2012.00353.x .
PMID 22928673 .
15. ^ "NEJM Journal Watch: Summaries of and
commentary on original medical and scientific articles
from key medical journals" . www.jwatch.org.
16. ^ "Preferred Usage - Emerging Infectious Disease
journal - CDC" . CDC.gov. Centers for Disease
Control and Prevention.
External links [ edit ]
3D structures of proteins from inner membranes of
Ellie Wyithe's gram-negative bacteria
V ·T ·E Microbiology: Bacteria [show]
V ·T ·E Prokaryotes: Bacteria [show]
classification (phyla and
orders)
Biology portal
Categories: Gram-negative bacteria Staining
Bacteriology
This page was last edited on 19 March 2021, at 19:17 (UTC).
Text is available under the Creative Commons Attribution-ShareAlike
License; additional terms may apply. By using this site, you agree to
the Terms of Use and Privacy Policy. Wikipedia® is a registered
trademark of the Wikimedia Foundation, Inc., a non-profit
organization.
Privacy policy About Wikipedia Disclaimers Contact Wikipedia
Mobile view Developers Statistics Cookie statement
:
You might also like
- Bacterial MorphologyDocument21 pagesBacterial MorphologyGaspar SantosNo ratings yet
- Bacterial Cell Structure: BacteriaDocument5 pagesBacterial Cell Structure: BacteriaHassan AljaberiNo ratings yet
- Lecture 1 Bacterial StructureDocument38 pagesLecture 1 Bacterial StructureAyat MostafaNo ratings yet
- Chapter 2 Pathogenic Bacteria_442be720c5ca44d60105704879d03d6fDocument33 pagesChapter 2 Pathogenic Bacteria_442be720c5ca44d60105704879d03d6fdaisysintszwaiNo ratings yet
- Del Rosario Microbio Activity 1 Bacteria CellDocument6 pagesDel Rosario Microbio Activity 1 Bacteria CellNico LokoNo ratings yet
- Bacterial Anatomy 2018Document79 pagesBacterial Anatomy 2018RocKyRiazNo ratings yet
- Infectious Disease NotesDocument43 pagesInfectious Disease NotessaguNo ratings yet
- 1 2020 PHARM232 - Intro To Bacteriology - NarratedDocument59 pages1 2020 PHARM232 - Intro To Bacteriology - Narrateddanst123No ratings yet
- Bacte in House ReviewDocument28 pagesBacte in House ReviewKenenth Jake BatiduanNo ratings yet
- USMLE Step 1 First Aid 2020 30th - MICROBIOLOGYDocument82 pagesUSMLE Step 1 First Aid 2020 30th - MICROBIOLOGYArt PuffNo ratings yet
- Structure & Functions of Prokaryotic Cell. Bacteriophages.: Lecturer: - Sergei V. RedkozubovDocument54 pagesStructure & Functions of Prokaryotic Cell. Bacteriophages.: Lecturer: - Sergei V. RedkozubovJustineNo ratings yet
- BDS Year 2 Lecture on Prokaryotic and Eukaryotic Cell StructuresDocument46 pagesBDS Year 2 Lecture on Prokaryotic and Eukaryotic Cell StructuresNarmathaa ThevarNo ratings yet
- Microbiology - FA (2017)Document83 pagesMicrobiology - FA (2017)asdasdasdasdasdNo ratings yet
- 1.3- Bacterial cell structure-MorphologyDocument39 pages1.3- Bacterial cell structure-Morphologyzekarias wondafrashNo ratings yet
- формы жизниDocument43 pagesформы жизниKhaled Mohammed GhalwashNo ratings yet
- Microbiology, Usmle EndpointDocument208 pagesMicrobiology, Usmle EndpointYazan M Abu-FaraNo ratings yet
- Struktur Dan Fungsi Sel Bakteri2Document41 pagesStruktur Dan Fungsi Sel Bakteri2Ois SariNo ratings yet
- StructureDocument15 pagesStructurefatimasanusimorikiNo ratings yet
- Endotoxin and Disease in Food AnimalsDocument8 pagesEndotoxin and Disease in Food Animalstaner_soysuren100% (1)
- Bacterial Structure 1 1 2Document41 pagesBacterial Structure 1 1 2Nour MohammedNo ratings yet
- SGL 2 (Structure of Bacterial Cell)Document32 pagesSGL 2 (Structure of Bacterial Cell)rekanshwan5No ratings yet
- Act. 3 Cytology PeraltaDocument18 pagesAct. 3 Cytology PeraltaCogie PeraltaNo ratings yet
- Prokaryotic CellDocument29 pagesProkaryotic CellSeshime Thyrone DavidsonNo ratings yet
- 3.pahs Structure of Bacterial Cells 2Document42 pages3.pahs Structure of Bacterial Cells 2Srijan BhattaraiNo ratings yet
- Micro para 1Document4 pagesMicro para 1Reselle EspirituNo ratings yet
- AP Biology Bacteria and VirusesDocument51 pagesAP Biology Bacteria and VirusesMichael TsaiNo ratings yet
- Lecture 3 - Bacterial Anatomy and PhysiologyDocument26 pagesLecture 3 - Bacterial Anatomy and Physiologyapi-370335280% (5)
- COMPARE STRUCTURE AND FUNCTION OF PROKARYOTIC AND EUKARYOTIC CELLSDocument107 pagesCOMPARE STRUCTURE AND FUNCTION OF PROKARYOTIC AND EUKARYOTIC CELLSYani-dha CidHotNo ratings yet
- Functional Anatomy of Prokaryotes and EukaryotesDocument6 pagesFunctional Anatomy of Prokaryotes and EukaryotesMollyNo ratings yet
- Bacteria and Bacterial Diseases Lecture NotesDocument8 pagesBacteria and Bacterial Diseases Lecture NotesPowell KitagwaNo ratings yet
- 2023 Antibiotics GAS LectureDocument48 pages2023 Antibiotics GAS Lectureгазиза смагуловаNo ratings yet
- FA - MicrobiologyDocument80 pagesFA - MicrobiologyMargaret GraceNo ratings yet
- Functions of BacteriaDocument4 pagesFunctions of BacteriaSister RislyNo ratings yet
- General Microbiology - Lecture 2Document33 pagesGeneral Microbiology - Lecture 2Almoatazbellah AbdallahNo ratings yet
- Micro DD AEDocument15 pagesMicro DD AEmostafaeldeeb2003No ratings yet
- Micro TestDocument8 pagesMicro TestMark Frederick100% (1)
- CH 2Document30 pagesCH 2Mohamed CabdullahiNo ratings yet
- The Structure of Bacterial CellDocument49 pagesThe Structure of Bacterial CellanaNo ratings yet
- QMS MicrobiologyDocument21 pagesQMS MicrobiologyDr-Shubhaneel NeogiNo ratings yet
- Bacterial Cell AnatomyDocument3 pagesBacterial Cell AnatomyTreshia SusonNo ratings yet
- Module 1 Microorganisms in Food and SpoilageDocument24 pagesModule 1 Microorganisms in Food and SpoilageSiddNo ratings yet
- Bacterial Structure 117Document34 pagesBacterial Structure 117Rahmah Khairunnisa QonitaNo ratings yet
- FAQ in MicrobiologyDocument117 pagesFAQ in MicrobiologyVenkatesh Arumugam100% (1)
- Biochimica Et Biophysica Acta: Richard M. Epand, Chelsea Walker, Raquel F. Epand, Nathan A. MagarveyDocument8 pagesBiochimica Et Biophysica Acta: Richard M. Epand, Chelsea Walker, Raquel F. Epand, Nathan A. MagarveyAgung RahmadaniNo ratings yet
- Microbiology Notes 2Document69 pagesMicrobiology Notes 2Marie Llanes100% (1)
- 3 MicrobiologyDocument82 pages3 MicrobiologyBrush LowdryNo ratings yet
- Microbiology Summaries 1-30 ReviewedDocument53 pagesMicrobiology Summaries 1-30 ReviewedyepNo ratings yet
- Cell Structure and Function of ProkaryotesDocument37 pagesCell Structure and Function of ProkaryotesShafira AnandaNo ratings yet
- Part 10 ProkaryoticDocument54 pagesPart 10 ProkaryoticLê Thanh HằngNo ratings yet
- BACT 211 Introduction to Clinical BacteriologyDocument21 pagesBACT 211 Introduction to Clinical BacteriologyFelicityNo ratings yet
- Bacterial Structure, Growth, and Metabolism ExplainedDocument77 pagesBacterial Structure, Growth, and Metabolism ExplainedNelle ReneiNo ratings yet
- The Bacterial Cell: Structures of Prokaryotes and Differences from EukaryotesDocument3 pagesThe Bacterial Cell: Structures of Prokaryotes and Differences from EukaryotesanojanNo ratings yet
- Lecture 2 - Bacterial Structure, Pathogenicity, Growth and MetabolismDocument21 pagesLecture 2 - Bacterial Structure, Pathogenicity, Growth and MetabolismGd HfNo ratings yet
- Microbial Taxonomy - System That Involves in The Organization, Classification, Naming orDocument5 pagesMicrobial Taxonomy - System That Involves in The Organization, Classification, Naming orRedelle Mae NiniNo ratings yet
- General Characteristics of Bacteria and MollicutesDocument13 pagesGeneral Characteristics of Bacteria and MollicutesPrincess Mehra0% (1)
- Microbiology Guide: Hallmarks of Life & Microorganism TypesDocument26 pagesMicrobiology Guide: Hallmarks of Life & Microorganism TypesChandan SikderNo ratings yet
- Morphological CharacteristicsDocument9 pagesMorphological CharacteristicsHabibur Rahaman0% (1)
- 1.3 Bacterial MorphologyDocument4 pages1.3 Bacterial MorphologyJosh Miguel BorromeoNo ratings yet
- characteristicsDocument5 pagescharacteristicssamvetNo ratings yet
- Biology for Students: The Only Biology Study Guide You'll Ever Need to Ace Your CourseFrom EverandBiology for Students: The Only Biology Study Guide You'll Ever Need to Ace Your CourseNo ratings yet
- Safari - Mar 9, 2022 at 11:34 PM 3Document1 pageSafari - Mar 9, 2022 at 11:34 PM 3Syimah UmarNo ratings yet
- Safari - Mar 9, 2022 at 11:41 PM 2Document1 pageSafari - Mar 9, 2022 at 11:41 PM 2Syimah UmarNo ratings yet
- Safari - Mar 9, 2022 at 11:41 PMDocument1 pageSafari - Mar 9, 2022 at 11:41 PMSyimah UmarNo ratings yet
- Long Case OrthopaedicDocument24 pagesLong Case OrthopaedicSyimah UmarNo ratings yet
- Safari - Mar 9, 2022 at 11:35 PMDocument1 pageSafari - Mar 9, 2022 at 11:35 PMSyimah UmarNo ratings yet
- Safari - Mar 9, 2022 at 11:34 PM 2Document1 pageSafari - Mar 9, 2022 at 11:34 PM 2Syimah UmarNo ratings yet
- Sydney Sweeney Made You Look: Celebrities & EntertainmentDocument1 pageSydney Sweeney Made You Look: Celebrities & EntertainmentSyimah UmarNo ratings yet
- 9 Things You Need To Know About The Chainsmokers: 9. They Claim Their Penises Are 17 Inches Tip To TipDocument1 page9 Things You Need To Know About The Chainsmokers: 9. They Claim Their Penises Are 17 Inches Tip To TipSyimah UmarNo ratings yet
- Top 6 search results for number 6Document1 pageTop 6 search results for number 6Syimah UmarNo ratings yet
- Apartment Essentials: LifestyleDocument1 pageApartment Essentials: LifestyleSyimah UmarNo ratings yet
- Safari - 19 Apr 2021 at 2:18 PMDocument1 pageSafari - 19 Apr 2021 at 2:18 PMSyimah UmarNo ratings yet
- Safari - Mar 9, 2022 at 11:34 PMDocument1 pageSafari - Mar 9, 2022 at 11:34 PMSyimah UmarNo ratings yet
- Hypokalemia and Hyperkalemia: Normal Value: 3.5-5 Meq/L Hypokalemia HyperkalemiaDocument7 pagesHypokalemia and Hyperkalemia: Normal Value: 3.5-5 Meq/L Hypokalemia HyperkalemiaSyimah UmarNo ratings yet
- People Also Ask: VideosDocument1 pagePeople Also Ask: VideosSyimah UmarNo ratings yet
- Lyrics: Can You Feel It? Can You Feel It? Can You Feel It? Can You Feel It?Document1 pageLyrics: Can You Feel It? Can You Feel It? Can You Feel It? Can You Feel It?Syimah UmarNo ratings yet
- HX Taking SurgeryDocument1 pageHX Taking SurgerySyimah UmarNo ratings yet
- Stoma (Medicine) : Gastrointestinal StomataDocument1 pageStoma (Medicine) : Gastrointestinal StomataSyimah UmarNo ratings yet
- Safari - 19 Apr 2021 at 2:19 PMDocument1 pageSafari - 19 Apr 2021 at 2:19 PMSyimah UmarNo ratings yet
- Escherichia Coli: Biology and BiochemistryDocument1 pageEscherichia Coli: Biology and BiochemistrySyimah UmarNo ratings yet
- Cell Membrane: HistoryDocument1 pageCell Membrane: HistorySyimah UmarNo ratings yet
- DDXDocument1 pageDDXSyimah UmarNo ratings yet
- Biology: EtymologyDocument1 pageBiology: EtymologySyimah UmarNo ratings yet
- Ligand: HistoryDocument1 pageLigand: HistorySyimah UmarNo ratings yet
- Anatomy: Connective TissueDocument1 pageAnatomy: Connective TissueSyimah UmarNo ratings yet
- Hepatobiliary: Cholelithiasis, Cholecystitis, Choledocolithiasis, CholangitisDocument1 pageHepatobiliary: Cholelithiasis, Cholecystitis, Choledocolithiasis, CholangitisSyimah UmarNo ratings yet
- Glycocalyx: in Vascular Endothelial TissueDocument1 pageGlycocalyx: in Vascular Endothelial TissueSyimah UmarNo ratings yet
- Plant Anatomy: Structural DivisionsDocument1 pagePlant Anatomy: Structural DivisionsSyimah UmarNo ratings yet
- Cell-Cell Recognition: FundamentalsDocument1 pageCell-Cell Recognition: FundamentalsSyimah UmarNo ratings yet
- Note 3 Dec 2019 at 1 - 50 - 26 PMDocument1 pageNote 3 Dec 2019 at 1 - 50 - 26 PMSyimah UmarNo ratings yet
- Post Harvest Maturity IndicesDocument3 pagesPost Harvest Maturity IndicesPanis Daniel G.No ratings yet
- Mastering English and Social StudiesDocument33 pagesMastering English and Social StudiesGalerya MarahuyoNo ratings yet
- Ecology Reading and ReviewDocument8 pagesEcology Reading and Reviewjohan preciadoNo ratings yet
- AREVIEWONHIVAIDSDocument6 pagesAREVIEWONHIVAIDSanthonybolteyNo ratings yet
- SOP Disc35Document3 pagesSOP Disc35MA Orejas RamosNo ratings yet
- 0097 Stage 5 Primary Science Scheme of Work Tcm142-595394Document69 pages0097 Stage 5 Primary Science Scheme of Work Tcm142-595394Romana Miriam OlivettiNo ratings yet
- NewbookchapteranthuriumDocument8 pagesNewbookchapteranthuriumTaashi Tiyon RuwanpathiranaNo ratings yet
- GastroenteritisDocument4 pagesGastroenteritisJj Cor-BanoNo ratings yet
- Control of A Robotic Arm Using ECGDocument48 pagesControl of A Robotic Arm Using ECGAnaNo ratings yet
- Cell Organelle Review Worksheet 14-15Document2 pagesCell Organelle Review Worksheet 14-15Kirsten Troupe100% (1)
- LIFELINE SBSG Practice HandbookDocument140 pagesLIFELINE SBSG Practice HandbookAmethyst CareyNo ratings yet
- Carlos Simon, Antonio Pellicer-Stem Cells in Human Reproduction_ Basic Science and Therapeutic Potential, 2nd Edition (Reproductive Medicine & Assisted Reproductive Techniques)-Informa Healthcare (200Document273 pagesCarlos Simon, Antonio Pellicer-Stem Cells in Human Reproduction_ Basic Science and Therapeutic Potential, 2nd Edition (Reproductive Medicine & Assisted Reproductive Techniques)-Informa Healthcare (200yasirhistoNo ratings yet
- ScienceDocument168 pagesScienceadi22-22No ratings yet
- WEF Building BlockchainsDocument37 pagesWEF Building BlockchainsvsrajkumarNo ratings yet
- Giant Cell Tumor - CompleteDocument35 pagesGiant Cell Tumor - CompletewildanmalikNo ratings yet
- Bonne Pratique D'hygiéne PoissonDocument147 pagesBonne Pratique D'hygiéne PoissonLaila HomeNo ratings yet
- The Biomedical Engineering Handbook: Second EditionDocument13 pagesThe Biomedical Engineering Handbook: Second EditionEng-Mugahed AlmansorNo ratings yet
- Mock Cat 3Document95 pagesMock Cat 3Sahil k0% (1)
- Biology BeedDocument2 pagesBiology BeedRj Nerf Monteverde CalasangNo ratings yet
- BotanyDocument28 pagesBotanyAnnabelle Phen LeeNo ratings yet
- Patient Centered Medicineresident ปี 1 พศ. 57 BenjapornDocument45 pagesPatient Centered Medicineresident ปี 1 พศ. 57 BenjapornPao RattanawanNo ratings yet
- Viton A401CDocument8 pagesViton A401Crainer100% (1)
- Discover MARDIDocument33 pagesDiscover MARDIMARDI Scribd75% (4)
- AllDocument490 pagesAllStarla Ellison100% (1)
- Biology of Tendon Injury: Healing, Modeling and Remodeling: P. Sharma and N. MaffulliDocument10 pagesBiology of Tendon Injury: Healing, Modeling and Remodeling: P. Sharma and N. MaffulliBrianna RyanNo ratings yet
- Levapor Carriers and Their Application For Wastewater TreatmentDocument29 pagesLevapor Carriers and Their Application For Wastewater TreatmentAmit Christian100% (1)
- How To Save Marine LifeDocument10 pagesHow To Save Marine Lifeyash salveNo ratings yet
- Classification of NF by DR MKBDocument33 pagesClassification of NF by DR MKBiloveusonNo ratings yet
- Approach To Skin Adnexal TumorsDocument20 pagesApproach To Skin Adnexal TumorsNirmalya Chakrabarti100% (1)
- Immunology Short Quiz With AnswersDocument6 pagesImmunology Short Quiz With AnswersPeter SewehaNo ratings yet