Professional Documents
Culture Documents
BMB 251 EXAM 2 Materials
Uploaded by
haiOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
BMB 251 EXAM 2 Materials
Uploaded by
haiCopyright:
Available Formats
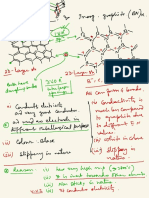
Protein
i. -
ligand binding -
non covalent bond hold them
together ( H / EAI VDWI
Hydrophobic)
a T water
✓
.
.
decreased e- interaction
by shielding charges →
unfavourable
as
b .
phosphatases → remove P
group off on
add P mechanism
kinase →
group
ON OFF
-
-
allosteric not active site
c .
→
{ -
regulation can
change enzyme conformation g wi crease binding ability (t)
inhibit C- )
enzyme
gene x pressures
d .
Enzymatic e a r confine to a
space enclosed
,
distinct membraneby / sub cell -
compartment
add C P)
regulation modification -
groups
destruction -
targeted proteolysis
bind to other Iunhibitors @ special
molecules
- activator
regulatory soles
2. GENES
Genomes complete set
of info
: a
-
3 B each diploid cells
of DNA
-
nu → 2M in → 20000 proteins
strands
-
Replication origins : a
leading
centromere and specialized DNA condensed
allows copy of each deeps
Sgc I
-
-
-
chromo → cell division
-
Telerosomes : repeated nuc sequences →
protect the end
of chromo
from being
(fate melt )
\ helps effectively
mistaken for Dna repair
replicate the ends
4 .
Chromatins
-
the complex og Dh AS Pro →
prevent entanglement
\
( regulate
↳ CD
reinforce DNA
during
prevent DNA
Damage
gene xpression s DNA replication
-
Euchromatin vs Heterochromatin → less vs more compacted
a . Histone : octamer = 2x ( H2 A , HAB , Hz , Ha)
wrapped around
"
particle
"
Nu histone octane 147 bp of beads
{
→ .
core = t DNA →
→
linker DNA = 20 -
so bp
↳ NUCLEOSOMES =
(core tlenker ) xn →
chromatin
> lysine t CHATS)
/
* Histone
modification →
acetylation of lysine activates transcription ↳ -
C HDACs)
I mono methylation activation euchre remove ⑦ lysine
[
→ t
dis tri → hetero t
repression to loosen chromatin
5 DNA
.
Rep '
3
- I
One polimerase @
s
Primer strand →
daughter strands
'
each strand
strand parent strand C 2 @ rep fork)
3
a .
Template →
Lagging strand → Okazaki ( 200 bp)
'
s
DNA primase synthesizes RNA primer
b. DNA pole proofreading mechanism
Accuracy : I mistake I 109 me
1 . Dna pole can
only tighten its
fingers s
form covalent bonds
with ( V) pairing
2 .
Exonucleolylic proof reading →
if previous one is Cx) → E cannot add new hue
→ cleaves the old ones
3. mismatch repair : →
newly synthesized lagging strand
transiently contains
↳
"
single -
strand break ( top I) aka
"
Nicks
Nuts mismatched base pair
{
t
Mut L scan
nearby DNA
for
nidseargely confined to
newly rep DNA → remove
error
selectively
6 . Bacterial Replication
roughly around chromosome
←
{
•
°
-
E coli
-
⇒
→
-
→ . : 4.6×10 me
pairs
→
- - - -
-
500 -
1000 nu / s ~ 40 mins
replicates theends sequence s→ tee
7 tease Rna contains a
templating DNA
-
¥nih9a%
repeats
several ④ protein domains → needed to assemble the E @ tneends
properly
strands need
④ The lagging RNA pole → @ the
very lip of a Centar DNA ,
there is no more peace
→ Euler have a special sequences called teeemers ( contains n -time tandem
GGGTTTA x thousands) →
repeat recognized by sequence specific DNA
binding
-
j
protein
You might also like
- IB Chemistry (Periodic Table)Document1 pageIB Chemistry (Periodic Table)hyunjinp0107No ratings yet
- Catetan Semester 5 Sisken EpDocument81 pagesCatetan Semester 5 Sisken EpAdzhani ZalfaNo ratings yet
- Colloquium 1Document8 pagesColloquium 1lorenzotolla.univNo ratings yet
- CytoskeletonDocument3 pagesCytoskeletonPASSORN SAE JEWNo ratings yet
- Ca MamaeDocument4 pagesCa MamaeAlvita Vania ANo ratings yet
- Dgcat Particular CategoryDocument1 pageDgcat Particular CategoryOlaNo ratings yet
- Energy and Momentum Live Class-4 Teacher NotesDocument19 pagesEnergy and Momentum Live Class-4 Teacher Notestarun singhNo ratings yet
- Student NotebookDocument3 pagesStudent NotebookMai SamirNo ratings yet
- Sapienza 1Document1 pageSapienza 1tuniaipartnerzyNo ratings yet
- Formulario Por Revisar PDFDocument1 pageFormulario Por Revisar PDFDanilo Malaver FonsecaNo ratings yet
- SF2 - Lecture-02 - Breathing and Exchange of Gases - NotesDocument7 pagesSF2 - Lecture-02 - Breathing and Exchange of Gases - Notesdisha shuklaNo ratings yet
- Digestion - 6Document11 pagesDigestion - 6aaravrshah14No ratings yet
- Asma dan Pneumonia: Pendekatan Terapi Berdasarkan ABCDE dan CURB-65Document5 pagesAsma dan Pneumonia: Pendekatan Terapi Berdasarkan ABCDE dan CURB-65iaw iawNo ratings yet
- Emi 2024Document6 pagesEmi 2024pingjin010No ratings yet
- Lect13 Weight Bal SlidesDocument4 pagesLect13 Weight Bal Slidesrashed44No ratings yet
- Differently: Piy0heaDocument1 pageDifferently: Piy0heaSweetthing777No ratings yet
- Lec 3 - Gauss's LawDocument8 pagesLec 3 - Gauss's LawFatemeh KeshavarzNo ratings yet
- HarshDocument20 pagesHarshHarsh Vashishtha67% (3)
- Atomic StructureDocument2 pagesAtomic StructureHems MadaviNo ratings yet
- Atomic Structure Mind MapDocument2 pagesAtomic Structure Mind Maplakshminivas PingaliNo ratings yet
- A-Level RDMS & KeysDocument18 pagesA-Level RDMS & KeysAsra BibiNo ratings yet
- PHARMACOLOGYDocument1 pagePHARMACOLOGYPublic AleeshaNo ratings yet
- JUNOS Cheat-Sheet Quick ReferenceDocument5 pagesJUNOS Cheat-Sheet Quick ReferencereagvafdvNo ratings yet
- Ancient IndiaDocument300 pagesAncient IndiaVIJAY PHOTOSTAT & PRINTOUTNo ratings yet
- Sunday 2pm ClassDocument14 pagesSunday 2pm ClassRajarshi MandalNo ratings yet
- Simple: EntersDocument4 pagesSimple: EntersScienceStudsNo ratings yet
- Chapter 2 DNA ReplicationDocument1 pageChapter 2 DNA ReplicationYuume YuuNo ratings yet
- Week 10 Unifiers and Most General UnifiersDocument6 pagesWeek 10 Unifiers and Most General Unifiersyallah.abibi.wahahaNo ratings yet
- Waves 2Document1 pageWaves 2fghhfgfNo ratings yet
- Basic Structure DoctrineDocument1 pageBasic Structure DoctrineKesava KumarNo ratings yet
- Skrypt Angielski ZimaDocument14 pagesSkrypt Angielski Zimamedrzyckiigorek2004No ratings yet
- Ch10-Fluids Sec 10.1-10.7Document6 pagesCh10-Fluids Sec 10.1-10.7basharrebhi29No ratings yet
- CATARACT - GLAUCOMADocument2 pagesCATARACT - GLAUCOMAggukNo ratings yet
- p07 PDFDocument1 pagep07 PDFRonal J Clavijo RNo ratings yet
- Wiring Diagram PDFDocument1 pageWiring Diagram PDFAlexander RojasNo ratings yet
- Physical and Chemical Classification of MatterDocument15 pagesPhysical and Chemical Classification of MatterKotyada ParthivNo ratings yet
- Hipoalb: Persona Corp YeenevgiaDocument2 pagesHipoalb: Persona Corp YeenevgiaJENNIFER DIANA MORENO PRECIADONo ratings yet
- Electricity PRV Years Final Shobhit NirwanDocument5 pagesElectricity PRV Years Final Shobhit NirwanRishabh JoshiNo ratings yet
- EMAT 251 Materials Science Chapter 6 Mechanics of MaterialsDocument40 pagesEMAT 251 Materials Science Chapter 6 Mechanics of MaterialsTaha Alper ŞenNo ratings yet
- 1 StoichiometryDocument4 pages1 StoichiometryKira BezkorovainaNo ratings yet
- bhramastra_revision_patrika_physics_electromagnetic_induction_mindmapsDocument2 pagesbhramastra_revision_patrika_physics_electromagnetic_induction_mindmapsyuvrajjadav882No ratings yet
- Electromagnetic Induction - Mind Map - Lakshya NEET 2024Document2 pagesElectromagnetic Induction - Mind Map - Lakshya NEET 2024Test UserNo ratings yet
- Chemistry 11 Short Notes of Structure of AtomDocument2 pagesChemistry 11 Short Notes of Structure of Atomkanit8713No ratings yet
- Macro Economic - LectureDocument7 pagesMacro Economic - LectureANYA AKBARNo ratings yet
- Act460-Scholar ch4 Part 1Document6 pagesAct460-Scholar ch4 Part 1Anfal KankouniNo ratings yet
- Class 12 Physics Derivations Shobhit NirwanDocument24 pagesClass 12 Physics Derivations Shobhit Nirwansuyash KumarNo ratings yet
- NAME/D1AaRAMDocument2 pagesNAME/D1AaRAMaliceNo ratings yet
- Untitled Notebook (2) - 1Document4 pagesUntitled Notebook (2) - 1poojaNo ratings yet
- 3 Ethics - Asp CoDocument4 pages3 Ethics - Asp Cosbracca1No ratings yet
- 19.R. Pancreas - Cai BiliareDocument10 pages19.R. Pancreas - Cai BiliareAna MîndrilăNo ratings yet
- Regression AnalysisDocument4 pagesRegression AnalysisThorn MailNo ratings yet
- Notes - Systems of ParticlesDocument9 pagesNotes - Systems of ParticlesGemaNo ratings yet
- Cell StainingDocument1 pageCell StainingAqiena BalqisNo ratings yet
- 2 PDFDocument1 page2 PDFSim Pei YingNo ratings yet
- UOM Electromagnetic Induction Notes 2Document5 pagesUOM Electromagnetic Induction Notes 2Arctic TurtleNo ratings yet
- Modul Bedah 2 - DR RianDocument18 pagesModul Bedah 2 - DR RianDendy AgusNo ratings yet
- 物理冶金2Document1 page物理冶金2游承翰No ratings yet
- Zruomponen K: KonfrasDocument12 pagesZruomponen K: KonfrasMuhammad Sobri MaulanaNo ratings yet
- The Pennsylvania State University: P A A T+a TDocument1 pageThe Pennsylvania State University: P A A T+a ThaiNo ratings yet
- HW#7Document2 pagesHW#7haiNo ratings yet
- Acetylene Production Rates and Membrane ReactorDocument2 pagesAcetylene Production Rates and Membrane ReactorhaiNo ratings yet
- HW 12Document2 pagesHW 12haiNo ratings yet
- HW#7Document2 pagesHW#7haiNo ratings yet
- HW Che 220Document3 pagesHW Che 220haiNo ratings yet
- HW 12 Che 220Document2 pagesHW 12 Che 220haiNo ratings yet
- Acetylene Production Rates and Membrane ReactorDocument2 pagesAcetylene Production Rates and Membrane ReactorhaiNo ratings yet
- ProteomicsDocument24 pagesProteomicswatson191No ratings yet
- Simple Color Tests For Amino Acids and Proteins LabDocument7 pagesSimple Color Tests For Amino Acids and Proteins Labpa3ciajp67% (3)
- Dr. Farzana Shahin Assistant Professor BioinformaticsDocument21 pagesDr. Farzana Shahin Assistant Professor BioinformaticsMuhammad BilalNo ratings yet
- Factors Controlling Growth 05-01-2020Document20 pagesFactors Controlling Growth 05-01-2020kashif manzoorNo ratings yet
- Morrissey 2004Document6 pagesMorrissey 2004Marija NikolicNo ratings yet
- Menegon2016 Ozonio e CancerDocument16 pagesMenegon2016 Ozonio e CancerJuju e Otávio FotosNo ratings yet
- AnticancerDocument78 pagesAnticancerRajkishor GogoiNo ratings yet
- 11 9700 23 2019 165347.inddDocument2 pages11 9700 23 2019 165347.inddAya TharwatNo ratings yet
- Molecular Regulation2Document24 pagesMolecular Regulation2Atharva PurohitNo ratings yet
- Glucose BCDocument17 pagesGlucose BCAditya ChawlaNo ratings yet
- Lecture Biochemistry of Connective TissueDocument97 pagesLecture Biochemistry of Connective TissueNeha FathimaNo ratings yet
- ProteinsDocument43 pagesProteinsBilalNo ratings yet
- Anatomy of Dermo-Epidermal Junction and Its Applied AspectDocument56 pagesAnatomy of Dermo-Epidermal Junction and Its Applied AspectSiddharth DashNo ratings yet
- SBIA031 Study Guide 2024Document10 pagesSBIA031 Study Guide 2024Itumeleng SefaraNo ratings yet
- Essay On ProteinDocument2 pagesEssay On ProteinLester MendigoriaNo ratings yet
- Basic Molecular Biology: Translation Process and Genetic Code DecodingDocument51 pagesBasic Molecular Biology: Translation Process and Genetic Code DecodingAleena MustafaNo ratings yet
- Protein Structure: Predictive Methods and Experimental MethodologiesDocument33 pagesProtein Structure: Predictive Methods and Experimental MethodologiesmarylaranjoNo ratings yet
- Organization and Evolution of The Nuclear Genome: Princess Lorraine Garcia Bs-Biology IvDocument25 pagesOrganization and Evolution of The Nuclear Genome: Princess Lorraine Garcia Bs-Biology IvLouella ArtatesNo ratings yet
- Problem Set 1 Introduction and Cell SignalingDocument9 pagesProblem Set 1 Introduction and Cell SignalingPreston Adhikari100% (1)
- McFadden and Sacco JMB 2021 UPDATEDDocument1 pageMcFadden and Sacco JMB 2021 UPDATEDNandan GokhaleNo ratings yet
- PDB 10,000th Entry: Bacteriophage phiX174 Virus StructureDocument3 pagesPDB 10,000th Entry: Bacteriophage phiX174 Virus StructureArul ManiNo ratings yet
- Chapter 18 - Apoptosis - 112612Document28 pagesChapter 18 - Apoptosis - 112612Anonymous nkR1PhIHNo ratings yet
- Calcium Signalling in Cancer-Sh ErbetDocument382 pagesCalcium Signalling in Cancer-Sh ErbetGreen ViktorNo ratings yet
- Gene Data - AG RedactedDocument153 pagesGene Data - AG RedactedAustin GomezNo ratings yet
- Lesson Plan March 29, 2019 Biochemistry Chapter 5 Topic: Enzymes Pp. 43-47Document3 pagesLesson Plan March 29, 2019 Biochemistry Chapter 5 Topic: Enzymes Pp. 43-47enilegnavemartinezNo ratings yet
- Alteration of Host Cell Behavior by PathogensDocument7 pagesAlteration of Host Cell Behavior by PathogensAbhijit SatpatiNo ratings yet
- Chapter 19 PDFDocument30 pagesChapter 19 PDFJeanPaule JoumaaNo ratings yet
- J. Gram, J. Jespersen (Auth.), J. Jespersen, R. M. Bertina, F. Haverkate (Eds.) - Laboratory Techniques in Thrombosis - A Manual-Springer Netherlands (1999)Document307 pagesJ. Gram, J. Jespersen (Auth.), J. Jespersen, R. M. Bertina, F. Haverkate (Eds.) - Laboratory Techniques in Thrombosis - A Manual-Springer Netherlands (1999)BipedalJoeNo ratings yet
- Molecular ClockworkDocument10 pagesMolecular Clockworkgarsa psikiatriNo ratings yet
- THE CELL Concept MapDocument3 pagesTHE CELL Concept MapDae PelayoNo ratings yet