Professional Documents
Culture Documents
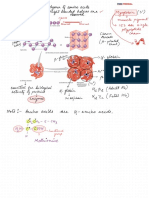
Individuals Level Filtering: Lmiss
Uploaded by
KS VelArcOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Individuals Level Filtering: Lmiss
Uploaded by
KS VelArcCopyright:
Available Formats
Individuals Level Filtering sexcheck
Framingham
Process 5
the binary file that contains the
NP1: step4
Data Import Check discordant sex raw genotype data
ped map txt
plink
Framingham Framingham fail-sex-qc
Process 6
—bfile Framingham NP1: step5
Genotype file SNP map file Individual with sex discrepancy
—check-sex Select Sex Problem Individual
—out Framingham Bash Shell
Process 9
Process 3
to calculate the mean homozygosity rate NP1: step9
NP1: step 3
Make BED file bed across X-chromosome markers for each Plot Heter vs. Missing txt
Framingham individual in the study fail-qc-inds
plink miss-vs-het.Rscript Process 13
the binary file that contains the NP1: step20 Failed individual; to be removed
—file raw-GWA-data Concentrate all failed individual
raw genotype data from consideration
log —make-bed Bash Shell
Framingham Process 7
—out raw-GWA-data
NP1: step7 Genotype and heterozygosity rate
Running log Process 14
imiss
plink NP1: step21
Framingham Removal of failed individual
—bfile Framingham
fam Missing genotype rate for plink
Framingham —missing (i)ndividual
Framingham.imiss-vs-het.pdf txt —file raw-GWA-data
.fam which is just the first six bim —out Framingham fail-imisshet-qc
—remove fail-qc-inds.txt
columns of .ped. Framingham lmiss
to calculate the missing data rates Failed individual; to be removed
Framingham —make-bed
a revsied map file .bim which from consideration
contains two extra columns that Missing genotype rate for SNP —out clean-inds-GWA-data
give the allele names for each
SNP Process 8
NP1: step8
Calculate heterozygosity rate
plink
het Process 10
—bfile Framingham Framingham NP1: step10
—het Heterozygosity rate Select genotype and heterozygosity
problems
—out Framingham
miss-vs-het-cutoff.Rscript
to calculate the outlying heterozygosity
rate
Process 16
NP1: step23 Plot histogram & Extract
failed marker txt
clean-ind-Hist.Rscript fail-lmiss-qc
imiss txt
SNP with excessing
clean-inds-Framingham fail-markers-qc
Process 15 missing data
bed NP1: step22 Missing genotype rate for markers with
Process 21
clean-inds-Framingham Calculate marker missing genotype (i)ndividual excessing missing
NP1: step25.1 data
Finalize marker filter
the binary file that contains the plink Process 17
Concentrate all failed marker
raw genotype data NP1: step23.2
—bfile clean-inds-Framingham
lmiss Extract Failed marker Bash Shell
—missing clean-inds-Framingham
Bash Shell
Process 22
—out clean-inds-Framingham Missing genotype rate for SNP
NP1: step26
Column QC Finalize
Process 19 Removal of failed marker
NP1: step25 plink
Process 18 Extract P < 0.0001
NP1: step24 txt
Calculate genotype call missing —file raw-GWA-data
Bash Shell fail-diffmiss-qc
rates between cases and control clean-inds-Framingham
—exclude fail-markers-qc.txt
Missing data rate
plink Call rates
between cases and —maf 0.01
—bfile clean-inds-Framingham controls
—geno 0.05
—test-missing —hwe 0.00001
—out clean-inds-Framingham —make-bed
—out clean-GWA-data
Marker Level Filtering
Statistical Analysis Process 23A1
NP2: step2 Result Interpretation
chi-square with PED default
assoc
plink
Framingham-result
—bfile clean-Framingham
Associate analysis results Process 25
—assoc NP1: step10
Result Annotation
—out Framingham-result
Result_Interpretation.Rscript
Process 23A2 assoc
NP2: step2 Framingham-adjusted-result
chi-square with PED default:
Multiple Comparison Correction Adjustment Associate analysis results
plink
—bfile clean-Framingham
bed assoc
clean-Framingham —assoc
covar-SEX
the binary file that contains the —out Framingham-result
Associate analysis results
genotype data
Process 24
Process 23B1 NP1: step10
NP2: step2.1
Plot Mahattan and QQ plots Data Visualization
chi-square with co-var file: SEX manhattan-plot.Rscript
plink
—bfile clean-Framingham assoc
covar-ASTHMA
—pheno Framingham-Asthma.cov
Associate analysis results
-- pheno-name SEX
-- out covar-SEX
-- assoc
Process 23B2
NP2: step2.1
chi-square with co-var file: ASTHMA
plink
model
—bfile clean-Framingham
Framingham-model
—pheno Framingham-Asthma.cov
Associate analysis results
-- pheno-name ASTHMA
-- out covar-ASTHMA
-- assoc
Process 23C
NP2: step4(A)
chi-square testing all model
plink
—bfile clean-Framingham
-- model
-- out Framingham-model
You might also like
- Penetration Testing ToolsDocument1 pagePenetration Testing ToolsJulián GonzálezNo ratings yet
- APH120 BRsDocument1 pageAPH120 BRsScott MullerNo ratings yet
- Transcription )Document5 pagesTranscription )Kotchaphan ChansawangNo ratings yet
- E-Sax Setup Menu: Reset Hammer List IncrementDocument2 pagesE-Sax Setup Menu: Reset Hammer List Incrementgustavo caicedoNo ratings yet
- In Vitro Generation Of: Immortalized CD34+Document3 pagesIn Vitro Generation Of: Immortalized CD34+Mind NiramindNo ratings yet
- SF2-Lecture-02 Biomolecules NotesDocument16 pagesSF2-Lecture-02 Biomolecules Notesdisha shuklaNo ratings yet
- SH Oct09 Competitive MarketsDocument2 pagesSH Oct09 Competitive MarketsSimply HiredNo ratings yet
- Note CompArchDocument2 pagesNote CompArchĐức MinhNo ratings yet
- Enumeration MindmapDocument1 pageEnumeration MindmapMohammad Sazzadur RahmanNo ratings yet
- Young2019 - Deconstructing The Sources of Genotype-Phenotype Associations in HumansDocument6 pagesYoung2019 - Deconstructing The Sources of Genotype-Phenotype Associations in HumansKS VelArcNo ratings yet
- Tutorial - Ernst - ChromHMMDocument55 pagesTutorial - Ernst - ChromHMMKS VelArcNo ratings yet
- The Long Walk To African GenomicsDocument3 pagesThe Long Walk To African GenomicsKS VelArcNo ratings yet
- Tutorial - Shane's Simple Guide To F-StatisticsDocument21 pagesTutorial - Shane's Simple Guide To F-StatisticsKS VelArcNo ratings yet
- Playing A Murder Mystery GameDocument1 pagePlaying A Murder Mystery GameKS VelArcNo ratings yet
- Name: Berdon, Beverly Bernadeth V. Section: BSOA 4-2Document3 pagesName: Berdon, Beverly Bernadeth V. Section: BSOA 4-2Bernadeth BerdonNo ratings yet
- 4 EdificiosDocument29 pages4 EdificiosRicardo AmadoNo ratings yet
- Innovation That Works Around You: Megaflex Dpa - The Best in Power ProtectionDocument16 pagesInnovation That Works Around You: Megaflex Dpa - The Best in Power ProtectionGeorgeNo ratings yet
- Academic Head S.B.W: Pakistan International School Doha QatarDocument6 pagesAcademic Head S.B.W: Pakistan International School Doha QatarGhulam Rasool MagsiNo ratings yet
- Murali Jampala - Senior .Net Full Stack DeveloperDocument8 pagesMurali Jampala - Senior .Net Full Stack Developeradityar117No ratings yet
- 15-Heat Stress PlanDocument2 pages15-Heat Stress PlanKhan MuhammadNo ratings yet
- XRT 2011 Aerated Emulsions GVD SkyScan LeuvenDocument14 pagesXRT 2011 Aerated Emulsions GVD SkyScan LeuvenClaudio Yamamoto MorassutiNo ratings yet
- Chapter 5Document52 pagesChapter 5Pratham SuhasiaNo ratings yet
- Koe-064 Object Oriented ProgrammingDocument2 pagesKoe-064 Object Oriented Programmingsaksham agarwal0% (1)
- Mos 6545-1 CRTC RecreatedDocument12 pagesMos 6545-1 CRTC RecreatedarcarliniNo ratings yet
- Enterprise Javabeans (Ejb 3.X) : Fahad R. GolraDocument56 pagesEnterprise Javabeans (Ejb 3.X) : Fahad R. GolraAdrian BuzasNo ratings yet
- Computer Guy Meme - Căutare GoogleDocument1 pageComputer Guy Meme - Căutare GoogleDaria ConstantinNo ratings yet
- Samsung NP270E5E Lampard-AMD INT Rev 1.0 SchematicDocument41 pagesSamsung NP270E5E Lampard-AMD INT Rev 1.0 SchematicAndré Frota Paiva100% (1)
- Cambridge International AS and A Level Biology Course Book CD-ROMDocument7 pagesCambridge International AS and A Level Biology Course Book CD-ROMJane W.CarterNo ratings yet
- ChatGPT-4 X Midjourney Training SheetDocument6 pagesChatGPT-4 X Midjourney Training SheetJack100% (2)
- Characteristics and Quality AttributesDocument29 pagesCharacteristics and Quality AttributeshariniNo ratings yet
- Composite Microservice and Future of MicroserviceDocument12 pagesComposite Microservice and Future of MicroserviceRV devNo ratings yet
- Abstract Blood DonationDocument3 pagesAbstract Blood DonationRushikesh TamkarNo ratings yet
- MM2 Linear Algebra 2011 I 1x2Document33 pagesMM2 Linear Algebra 2011 I 1x2Aaron LeeNo ratings yet
- Symmetrix Cloning CommandsDocument2 pagesSymmetrix Cloning Commandsarun401No ratings yet
- P01696-1004-4 - 03 As Built Bill of MaterialsDocument15 pagesP01696-1004-4 - 03 As Built Bill of MaterialsMahmoud GaberNo ratings yet
- Makkar Reading 2019 EditionDocument181 pagesMakkar Reading 2019 EditionMunagala Sai SumanthNo ratings yet
- Stock Management System in PHP With Source Code - VIDEO - 2021Document9 pagesStock Management System in PHP With Source Code - VIDEO - 2021Abdulazeez Rabo100% (1)
- A.pt - Er': ManualDocument5 pagesA.pt - Er': ManualHenok DireNo ratings yet
- Deep Physical Neural Networks Trained With Backpropagation. Nature 2022, P L McmohanDocument11 pagesDeep Physical Neural Networks Trained With Backpropagation. Nature 2022, P L McmohanmahadimasnadNo ratings yet
- SKD 31&32 BSMRMU Numerical SimulationDocument25 pagesSKD 31&32 BSMRMU Numerical SimulationSudipta Roy RimoNo ratings yet
- RM Assignment - Calling Up AttendanceDocument3 pagesRM Assignment - Calling Up AttendancepresbanNo ratings yet
- QRCode-v-1 3Document32 pagesQRCode-v-1 3Mohammed KhalidNo ratings yet
- AMBCO 2500 ManualDocument22 pagesAMBCO 2500 ManualAlexandra JanicNo ratings yet
- Technical Solution 5G v2.4Document42 pagesTechnical Solution 5G v2.4alberto80% (5)