Professional Documents
Culture Documents
Reactome 2022 Table
Uploaded by
Backup Isa0 ratings0% found this document useful (0 votes)
16 views6 pagesteste
Original Title
Reactome_2022_table
Copyright
© © All Rights Reserved
Available Formats
TXT, PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this Documentteste
Copyright:
© All Rights Reserved
Available Formats
Download as TXT, PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
16 views6 pagesReactome 2022 Table
Uploaded by
Backup Isateste
Copyright:
© All Rights Reserved
Available Formats
Download as TXT, PDF, TXT or read online from Scribd
You are on page 1of 6
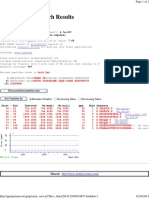
Term Overlap P-value Adjusted P-value Old P-value Old Adjusted P-value
Odds Ratio Combined Score Genes
Cytoprotection By HMOX1 R-HSA-9707564 2/57 0.004985271935017297
0.2881227220718465 0 0 20.683636363636364 109.64948564569971
COX8A;BRIP1
Regulation Of HMOX1 Expression And Activity R-HSA-9707587 1/5
0.009216658544821541 0.2881227220718465 0 0 138.60416666666666
649.6020692107373 BRIP1
Maturation Of Protein 3A R-HSA-9683673 1/9 0.016530465500484762
0.2881227220718465 0 0 69.28819444444444 284.25829643277825
ST6GALNAC4
Regulation Of BACH1 Activity R-HSA-9708530 1/11 0.020167646885642113
0.2881227220718465 0 0 55.425 216.36122001333428 BRIP1
Cytosolic Iron-Sulfur Cluster Assembly R-HSA-2564830 1/12 0.02198132687986945
0.2881227220718465 0 0 50.38383838383838 192.34342502593887
BRIP1
Tandem Pore Domain Potassium Channels R-HSA-1296346 1/12 0.02198132687986945
0.2881227220718465 0 0 50.38383838383838 192.34342502593887
KCNK4
MET Activates PTK2 Signaling R-HSA-8874081 1/17 0.031000847034435945
0.2881227220718465 0 0 34.630208333333336 120.29636590931148
LAMA5
Phase 4 - Resting Membrane Potential R-HSA-5576886 1/19 0.03458595035428571
0.2881227220718465 0 0 30.77932098765432 103.55110778477112
KCNK4
Initiation Of Nuclear Envelope (NE) Reformation R-HSA-2995383 1/19
0.03458595035428571 0.2881227220718465 0 0 30.77932098765432
103.55110778477112 ANKLE2
Laminin Interactions R-HSA-3000157 1/22 0.03993942012916417 0.2881227220718465
0 0 26.37830687830688 84.948474454672 LAMA5
RHOBTB1 GTPase Cycle R-HSA-9013422 1/23 0.04171748319247683 0.2881227220718465
0 0 25.178030303030305 79.98644731441095 RBBP6
Impaired BRCA2 Binding To PALB2 R-HSA-9709603 1/24 0.04349234219507745
0.2881227220718465 0 0 24.082125603864736 75.50156731119475
BRIP1
Cellular Response To Chemical Stress R-HSA-9711123 2/182 0.044548578404360946
0.2881227220718465 0 0 6.280317460317461 19.539166898746526
COX8A;BRIP1
Defective HDR Thru Homologous Recombination (HRR) Due To BRCA1 Loss-Of-Function R-
HSA-9701192 1/25 0.04526400273569332 0.2881227220718465 0 0
23.077546296296298 71.43061833824453 BRIP1
Termination Of O-glycan Biosynthesis R-HSA-977068 1/25 0.04526400273569332
0.2881227220718465 0 0 23.077546296296298 71.43061833824453
ST6GALNAC4
RHO GTPase Cycle R-HSA-9012999 3/441 0.04761262320650789 0.2881227220718465
0 0 3.9333199033037873 11.975611390815642 ANKLE2;DOCK8;RBBP6
Resolution Of D-loop Structures Thru Synthesis-Dependent Strand Annealing (SDSA) R-
HSA-5693554 1/27 0.048797750803848496 0.2881227220718465 0 0
21.300213675213676 64.32815883418561 BRIP1
MET Promotes Cell Motility R-HSA-8875878 1/28 0.050559849504957594
0.2881227220718465 0 0 20.51028806584362 61.214954602737194
LAMA5
Translation Of Structural Proteins R-HSA-9683701 1/30 0.05407452407786471
0.2881227220718465 0 0 19.093869731800766 55.70430487305193
ST6GALNAC4
Nuclear Signaling By ERBB4 R-HSA-1251985 1/32 0.05757653857644631
0.2881227220718465 0 0 17.86021505376344 50.984486277080904
ADAP1
Resolution Of D-loop Structures Thru Holliday Junction Intermediates R-HSA-5693568
1/33 0.05932281215138037 0.2881227220718465 0 0
17.30121527777778 48.8718043349592 BRIP1
Sialic Acid Metabolism R-HSA-4085001 1/33 0.05932281215138037
0.2881227220718465 0 0 17.30121527777778 48.8718043349592
ST6GALNAC4
Synthesis Of Bile Acids And Bile Salts R-HSA-192105 1/34 0.061065937308456206
0.2881227220718465 0 0 16.776094276094277 46.90262214500739
OSBPL3
Resolution Of D-Loop Structures R-HSA-5693537 1/34 0.061065937308456206
0.2881227220718465 0 0 16.776094276094277 46.90262214500739
BRIP1
RHOBTB GTPase Cycle R-HSA-9706574 1/35 0.06280591956350945 0.2881227220718465
0 0 16.28186274509804 45.063408386674496 RBBP6
Impaired BRCA2 Binding To RAD51 R-HSA-9709570 1/35 0.06280591956350945
0.2881227220718465 0 0 16.28186274509804 45.063408386674496
BRIP1
HDR Thru Single Strand Annealing (SSA) R-HSA-5685938 1/37 0.06627647739373588
0.2881227220718465 0 0 15.375771604938272 41.72861769022075
BRIP1
Presynaptic Phase Of Homologous DNA Pairing And Strand Exchange R-HSA-5693616
1/40 0.07145887974209389 0.2881227220718465 0 0
14.190883190883191 37.444534151430766 BRIP1
Intraflagellar Transport R-HSA-5620924 1/41 0.07318011991211305
0.2881227220718465 0 0 13.835416666666667 36.177283050000156
DYNC2H1
Defective Homologous Recombination Repair (HRR) Due To BRCA2 Loss Of Function R-
HSA-9701190 1/41 0.07318011991211305 0.2881227220718465 0 0
13.835416666666667 36.177283050000156 BRIP1
Non-integrin membrane-ECM Interactions R-HSA-3000171 1/41 0.07318011991211305
0.2881227220718465 0 0 13.835416666666667 36.177283050000156
LAMA5
Homologous DNA Pairing And Strand Exchange R-HSA-5693579 1/43
0.07661329204429566 0.2881227220718465 0 0 13.17526455026455
33.84705294145367 BRIP1
Glycosphingolipid Metabolism R-HSA-1660662 1/45 0.08003408946440181
0.2881227220718465 0 0 12.575126262626263 31.755999257171126
NAT8
Bile Acid And Bile Salt Metabolism R-HSA-194068 1/45 0.08003408946440181
0.2881227220718465 0 0 12.575126262626263 31.755999257171126
OSBPL3
Heme Signaling R-HSA-9707616 1/45 0.08003408946440181 0.2881227220718465
0 0 12.575126262626263 31.755999257171126 BRIP1
Diseases Of DNA Repair R-HSA-9675135 1/51 0.09022266636383927
0.29303629305574436 0 0 11.062777777777777 26.611230877061317
BRIP1
SARS-CoV-1 Infection R-HSA-9678108 1/52 0.09191005432406149
0.29303629305574436 0 0 10.845315904139433 25.887170950029557
ST6GALNAC4
RHOJ GTPase Cycle R-HSA-9013409 1/54 0.09527570001266539
0.29303629305574436 0 0 10.435010482180294 24.532506003993255
DOCK8
ECM Proteoglycans R-HSA-3000178 1/55 0.09695396842829543
0.29303629305574436 0 0 10.241255144032921 23.89816310810052
LAMA5
Signaling By ERBB4 R-HSA-1236394 1/57 0.10030142301459057
0.29303629305574436 0 0 9.874503968253968 22.7071663783518 ADAP1
Extracellular Matrix Organization R-HSA-1474244 2/291 0.10083284288854989
0.29303629305574436 0 0 3.890064260998517 8.924940022914297
LAMA5;P4HA3
Late SARS-CoV-2 Infection Events R-HSA-9772573 1/58 0.10197061980620958
0.29303629305574436 0 0 9.700779727095517 22.14756449052859
ST6GALNAC4
O-linked Glycosylation Of Mucins R-HSA-913709 1/59 0.10363680333472576
0.29303629305574436 0 0 9.533045977011493 21.61010698803553
ST6GALNAC4
RAB Geranylgeranylation R-HSA-8873719 1/62 0.10861732713979637
0.29303629305574436 0 0 9.062841530054644 20.11882244883129 RAB25
Synthesis Of Substrates In N-glycan Biosythesis R-HSA-446219 1/63
0.1102715103728266 0.29303629305574436 0 0 8.91621863799283
19.658565146558267 ST6GALNAC4
Signaling By MET R-HSA-6806834 1/63 0.1102715103728266
0.29303629305574436 0 0 8.91621863799283 19.658565146558267
LAMA5
Signaling By Rho GTPases R-HSA-194315 3/644 0.1155842236929177
0.29303629305574436 0 0 2.659722859502615 5.7390324405857704
ANKLE2;DOCK8;RBBP6
Collagen Biosynthesis And Modifying Enzymes R-HSA-1650814 1/67
0.11685842652812153 0.29303629305574436 0 0 8.37415824915825
17.977576828172595 P4HA3
HDR Thru Homologous Recombination (HRR) R-HSA-5685942 1/67 0.11685842652812153
0.29303629305574436 0 0 8.37415824915825 17.977576828172595
BRIP1
Signaling By Rho GTPases, Miro GTPases And RHOBTB3 R-HSA-9716542 3/660
0.12200836008936249 0.29303629305574436 0 0 2.592801504163309
5.454387620447978 ANKLE2;DOCK8;RBBP6
RHOG GTPase Cycle R-HSA-9013408 1/74 0.12827153849910913
0.29303629305574436 0 0 7.568493150684931 15.542701942518896
ANKLE2
Nuclear Envelope (NE) Reassembly R-HSA-2995410 1/75 0.1298902228088851
0.29303629305574436 0 0 7.4658408408408405 15.238271103442942
ANKLE2
SUMOylation Of DNA Damage Response And Repair Proteins R-HSA-3108214 1/76
0.13150598255242812 0.29303629305574436 0 0 7.365925925925926
14.943275535371535 SMC5
G2/M DNA Damage Checkpoint R-HSA-69473 1/77 0.13311882283696222
0.29303629305574436 0 0 7.268640350877193 14.657308812510841
BRIP1
Biosynthesis Of N-glycan Precursor (Dolichol LLO) And Transfer To Protein R-HSA-
446193 1/77 0.13311882283696222 0.29303629305574436 0 0
7.268640350877193 14.657308812510841 ST6GALNAC4
Nuclear Events Mediated By NFE2L2 R-HSA-9759194 1/78 0.1347287487922272
0.29303629305574436 0 0 7.173881673881674 14.379986917991172
BRIP1
Amyloid Fiber Formation R-HSA-977225 1/79 0.1363357655606339
0.29303629305574436 0 0 7.081552706552706 14.110946742753484
NAT8
Processing Of DNA Double-Strand Break Ends R-HSA-5693607 1/80
0.13793987823465348 0.29303629305574436 0 0 6.991561181434599
13.849844705127882 BRIP1
Transcriptional Regulation By TP53 R-HSA-3700989 2/354 0.1391072355566644
0.29303629305574436 0 0 3.183603896103896 6.2796910446900345
COX8A;BRIP1
TP53 Regulates Metabolic Genes R-HSA-5628897 1/81 0.13954109193130684
0.29303629305574436 0 0 6.903819444444444 13.596355470642333
COX8A
RAC2 GTPase Cycle R-HSA-9013404 1/87 0.14908777997199146
0.29817880283959913 0 0 6.420219638242894 12.219090543005988
ANKLE2
Sphingolipid Metabolism R-HSA-428157 1/89 0.15224704731157518
0.29817880283959913 0 0 6.273674242424242 11.808628150096421
NAT8
Collagen Formation R-HSA-1474290 1/90 0.1538223982902694
0.29817880283959913 0 0 6.202871410736579 11.611506075891745
P4HA3
Regulation Of TP53 Activity Thru Phosphorylation R-HSA-6804756 1/90
0.1538223982902694 0.29817880283959913 0 0 6.202871410736579
11.611506075891745 BRIP1
Respiratory Electron Transport R-HSA-611105 1/90 0.1538223982902694
0.29817880283959913 0 0 6.202871410736579 11.611506075891745
COX8A
Expression And Translocation Of Olfactory Receptors R-HSA-9752946 2/393
0.16419132696725436 0.3094089224569546 0 0 2.8603580562659845
5.167874411866651 OR4N5;OR5A1
Hedgehog Off State R-HSA-5610787 1/98 0.1663230817512098 0.3094089224569546
0 0 5.689003436426117 10.205065817539719 DYNC2H1
KEAP1-NFE2L2 Pathway R-HSA-9755511 1/100 0.16942006718012562 0.3094089224569546
0 0 5.57351290684624 9.895070145641316 BRIP1
Olfactory Signaling Pathway R-HSA-381753 2/401 0.16943821944071324
0.3094089224569546 0 0 2.801861797350519 4.974052523121619
OR4N5;OR5A1
Potassium Channels R-HSA-1296071 1/102 0.1725058575997924 0.3105105436796263
0 0 5.462596259625963 9.599551979063234 KCNK4
Interleukin-4 And Interleukin-13 Signaling R-HSA-6785807 1/107
0.18017161336794374 0.31530032339390157 0 0 5.203616352201258
8.918194337277319 LAMA5
O-linked Glycosylation R-HSA-5173105 1/107 0.18017161336794374
0.31530032339390157 0 0 5.203616352201258 8.918194337277319
ST6GALNAC4
Degradation Of Extracellular Matrix R-HSA-1474228 1/109 0.18321853019841125
0.3162402028082167 0 0 5.106738683127572 8.666522046159436 LAMA5
Respiratory Electron Transport, ATP Synthesis By Chemiosmotic Coupling, Heat
Production By Uncoupling Proteins R-HSA-163200 1/112 0.18776825697049568
0.31971351862543856 0 0 4.967967967967968 8.309158689911323 COX8A
HDR Thru Homologous Recombination (HRR) Or Single Strand Annealing (SSA) R-HSA-
5693567 1/114 0.19078769890816324 0.3205233341657142 0 0
4.8795476892822025 8.083429391488162 BRIP1
Homology Directed Repair R-HSA-5693538 1/119 0.1982886022832236
0.3287416301011339 0 0 4.671610169491525 7.558813448212846 BRIP1
Cardiac Conduction R-HSA-5576891 1/126 0.20867632029573777
0.34147034230211637 0 0 4.408444444444444 6.90790430957761 KCNK4
Cell Surface Interactions At Vascular Wall R-HSA-202733 1/134
0.22038768224462837 0.3516802840690093 0 0 4.141604010025063
6.263625610568688 SELE
Signaling By Hedgehog R-HSA-5358351 1/135 0.22183969142755683 0.3516802840690093
0 0 4.1104892205638475 6.18957577258348 DYNC2H1
Factors Involved In Megakaryocyte Development And Platelet Production R-HSA-983231
1/136 0.22328906925016465 0.3516802840690093 0 0
4.079835390946502 6.116848538578219 DOCK8
Signaling By Receptor Tyrosine Kinases R-HSA-9006934 2/496 0.2335233451451768
0.36284043641189767 0 0 2.2520532099479467 3.2755511011402496
LAMA5;ADAP1
G2/M Checkpoints R-HSA-69481 1/148 0.24047803533135842 0.36284043641189767
0 0 3.744520030234316 5.33641481020768 BRIP1
DNA Double-Strand Break Repair R-HSA-5693532 1/149 0.24189362427459846
0.36284043641189767 0 0 3.7190315315315314 5.278262347243474
BRIP1
CDC42 GTPase Cycle R-HSA-9013148 1/149 0.24189362427459846
0.36284043641189767 0 0 3.7190315315315314 5.278262347243474
DOCK8
Metabolism Of Steroids R-HSA-8957322 1/153 0.24753035347454788
0.36692734750344747 0 0 3.6204312865497075 5.054926044171352
OSBPL3
Regulation Of TP53 Activity R-HSA-5633007 1/157 0.25312629653250535
0.3708594577104148 0 0 3.5268874643874644 4.845473309088604
BRIP1
Citric Acid (TCA) Cycle And Respiratory Electron Transport R-HSA-1428517 1/163
0.2614443643251304 0.37864356212605094 0 0 3.395233196159122
4.55482000338841 COX8A
SUMO E3 Ligases SUMOylate Target Proteins R-HSA-3108232 1/168 0.2683071722844291
0.3841670875890689 0 0 3.2927478376580175 4.332014095552551
SMC5
SUMOylation R-HSA-2990846 1/174 0.27646066709821937 0.391393753419951 0
0 3.1775850995504173 4.085378967174108 SMC5
RAC1 GTPase Cycle R-HSA-9013149 1/178 0.2818471457810456 0.3945860040934638
0 0 3.105147520401758 3.93232898320769 DOCK8
Hemostasis R-HSA-109582 2/576 0.28869909865932475 0.3997372135282958 0
0 1.930214036834246 2.398040619771884 DOCK8;SELE
Cilium Assembly R-HSA-5617833 1/186 0.2925033047389258 0.40060235214244183
0 0 2.9696696696696696 3.6505534946983973 DYNC2H1
Signal Transduction R-HSA-162582 6/2465 0.30188921018496356
0.4090111879925313 0 0 1.3777433785042439 1.6501166073028377
DYNC2H1;LAMA5;ANKLE2;DOCK8;RBBP6;ADAP1
Muscle Contraction R-HSA-397014 1/196 0.30560732477573394 0.4096438608696008
0 0 2.815954415954416 3.3381851399708733 KCNK4
Sensory Perception R-HSA-9709957 2/616 0.31625406306408266 0.4194527573270991
0 0 1.8007445323406235 2.073034017123773 OR4N5;OR5A1
Cell Cycle R-HSA-1640170 2/654 0.34224858743298225 0.4492012710057892
0 0 1.6924627519719544 1.8146889290307093 ANKLE2;BRIP1
Mitotic Anaphase R-HSA-68882 1/232 0.3508550349420652 0.45266148675240614
0 0 2.3727753727753726 2.485202563550029 ANKLE2
Mitotic Metaphase And Anaphase R-HSA-2555396 1/233 0.3520700452518714
0.45266148675240614 0 0 2.3624281609195403 2.466198127336593
ANKLE2
Cellular Responses To Stress R-HSA-2262752 2/722 0.3880251880319014
0.49072700676413256 0 0 1.5272222222222223 1.4457984058808009
COX8A;BRIP1
Metabolism Of Lipids R-HSA-556833 2/732 0.39465518026129476
0.49072700676413256 0 0 1.505518590998043 1.3997451556057046
NAT8;OSBPL3
Cell Cycle Checkpoints R-HSA-69620 1/271 0.3966360137886954
0.49072700676413256 0 0 2.026028806584362 1.8735423026583014
BRIP1
Cellular Responses To Stimuli R-HSA-8953897 2/736 0.3972988879611068
0.49072700676413256 0 0 1.4970027247956403 1.381832938655755
COX8A;BRIP1
Organelle Biogenesis And Maintenance R-HSA-1852241 1/275 0.4011498547357592
0.49072700676413256 0 0 1.9960462287104623 1.823228983167314
DYNC2H1
Asparagine N-linked Glycosylation R-HSA-446203 1/282 0.40897014061580184
0.49209501754739543 0 0 1.9456306840648478 1.7396139435427633
ST6GALNAC4
SARS-CoV-2 Infection R-HSA-9694516 1/283 0.4100791812894962
0.49209501754739543 0 0 1.938632781717888 1.7281069796670696
ST6GALNAC4
Antigen Processing: Ubiquitination And Proteasome Degradation R-HSA-983168 1/307
0.4360968691519418 0.5172706346733 0 0 1.7844045025417574
1.480861028880145 RBBP6
DNA Repair R-HSA-73894 1/310 0.4392695072225643 0.5172706346733 0 0
1.7668104998202085 1.453452775148989 BRIP1
Metabolism Of Proteins R-HSA-392499 4/1890 0.4679828024853053
0.5459799362328562 0 0 1.1617982582987885 0.8821809876564501
NAT8;RAB25;SMC5;ST6GALNAC4
Post-translational Protein Modification R-HSA-597592 3/1383 0.4762944769880739
0.5505789367018102 0 0 1.1881713554987212 0.8812892303013922
RAB25;SMC5;ST6GALNAC4
SARS-CoV Infections R-HSA-9679506 1/369 0.49824901992605286 0.5707216046425697
0 0 1.4790911835748792 1.0304166928796366 ST6GALNAC4
Class I MHC Mediated Antigen Processing And Presentation R-HSA-983169 1/378
0.5066980091430833 0.5721262612294081 0 0 1.4431181844974947
0.9810896044709914 RBBP6
M Phase R-HSA-68886 1/380 0.5085566766483628 0.5721262612294081 0
0 1.4353562005277045 0.9705571622535356 ANKLE2
Neuronal System R-HSA-112316 1/386 0.5140918888214097 0.5732352034645807
0 0 1.4125541125541126 0.9398474801528096 KCNK4
Metabolism R-HSA-1430728 4/2049 0.5335230324226847 0.5896833516250727
0 0 1.062043417055642 0.6672320020540996 COX8A;BRIP1;NAT8;OSBPL3
Signaling By Interleukins R-HSA-449147 1/453 0.5719253232314184
0.6266312237144236 0 0 1.1990535889872174 0.669967415727626
LAMA5
Neutrophil Degranulation R-HSA-6798695 1/468 0.5839241652598034
0.6342624553684072 0 0 1.1596478705686415 0.6238721839118695
NAT8
Cell Cycle, Mitotic R-HSA-69278 1/523 0.6251835292024217 0.6732745699103002
0 0 1.0345359727543635 0.4859319182751486 ANKLE2
Generic Transcription Pathway R-HSA-212436 2/1190 0.655021956921399
0.6994302251872565 0 0 0.903078403078403 0.38208030055362024
COX8A;BRIP1
RNA Polymerase II Transcription R-HSA-73857 2/1312 0.7080436257060957
0.7465145951657229 0 0 0.8136532170119957 0.2809134224545137
COX8A;BRIP1
Immune System R-HSA-168256 3/1943 0.7109662811102123 0.7465145951657229
0 0 0.8197240751970891 0.27963269911604577 LAMA5;NAT8;RBBP6
Cytokine Signaling In Immune System R-HSA-1280215 1/702 0.7337312609378602
0.7640507345303339 0 0 0.7632746869551434
0.23631934350168943 LAMA5
Adaptive Immune System R-HSA-1280218 1/733 0.7491219848220599
0.7736833613736029 0 0 0.7297738312082575
0.21079768532109525 RBBP6
Gene Expression (Transcription) R-HSA-74160 2/1449 0.7596009521251736
0.7781278046160316 0 0 0.7312074242274657 0.2010542898465067
COX8A;BRIP1
Infectious Disease R-HSA-5663205 1/961 0.8385658455119323 0.8500458113273357
0 0 0.5498553240740741 0.09680872322492648 ST6GALNAC4
Disease R-HSA-1643685 2/1736 0.843299415999341 0.8500458113273357 0
0 0.6007249958807053 0.10238348632353438 BRIP1;ST6GALNAC4
Innate Immune System R-HSA-168249 1/1035 0.860247641093035 0.860247641093035
0 0 0.5085160111755856 0.07654944575469012 NAT8
You might also like
- WikiPathway 2021 Human TableDocument2 pagesWikiPathway 2021 Human TableBackup IsaNo ratings yet
- Practical Methods for Biocatalysis and Biotransformations 2From EverandPractical Methods for Biocatalysis and Biotransformations 2John WhittallNo ratings yet
- Perseues26 07 17Document1,813 pagesPerseues26 07 17mayankgiaNo ratings yet
- On-line LC-NMR and Related TechniquesFrom EverandOn-line LC-NMR and Related TechniquesKlaus AlbertNo ratings yet
- Pattern Comp All CDB Nci60 Exp TERT MoldataDocument3,954 pagesPattern Comp All CDB Nci60 Exp TERT MoldataDustin MatozzoNo ratings yet
- 29599Document908 pages29599sunny bhatiaNo ratings yet
- A549 - HEK293 - Wt-MEFs - Copy Numbers - DDA - 230920Document2,183 pagesA549 - HEK293 - Wt-MEFs - Copy Numbers - DDA - 230920Tom SnellingNo ratings yet
- WB Protein ChecklistDocument9 pagesWB Protein ChecklistjazeserolfNo ratings yet
- Pmwasabi PaxillinDocument4 pagesPmwasabi PaxillinAlleleBiotechNo ratings yet
- Inhibitors of Signaling PathwaysDocument50 pagesInhibitors of Signaling PathwaysDvrm Mslm GzrNo ratings yet
- DBDGST Seq in C+Document1 pageDBDGST Seq in C+Anil KumarNo ratings yet
- Presentation 1Document6 pagesPresentation 1Rushank ShuklaNo ratings yet
- Supplemental Table 5Document3,410 pagesSupplemental Table 5dharmender singhNo ratings yet
- NO NTPN NTB Akun Jumlah Setor HasilDocument6 pagesNO NTPN NTB Akun Jumlah Setor HasilWahyu SuciatiNo ratings yet
- Data Barang Kasir KTP FM 2019Document50 pagesData Barang Kasir KTP FM 2019BoerhanNo ratings yet
- Suplementos Mass Spectofotometry 02Document5 pagesSuplementos Mass Spectofotometry 02danielaegomNo ratings yet
- 2G Hourly DailyDocument80 pages2G Hourly DailyRamane kakaNo ratings yet
- Kegg ResultDocument21 pagesKegg ResultSasikala RajendranNo ratings yet
- Ppat 1007974 s018Document701 pagesPpat 1007974 s018Navaneeth ReddyNo ratings yet
- All Ustrious Pmwasabi AnnexinDocument4 pagesAll Ustrious Pmwasabi AnnexinAlleleBiotechNo ratings yet
- Annual Consumption - Year 2011-12: Tender NameDocument4 pagesAnnual Consumption - Year 2011-12: Tender NameAkshay PatelNo ratings yet
- Nqx013 SuppDocument4 pagesNqx013 SuppCamilofonoNo ratings yet
- pmTFP1 PeroxisomesDocument4 pagespmTFP1 PeroxisomesAlleleBiotechNo ratings yet
- Answer KeysDocument149 pagesAnswer KeysJudeLaxNo ratings yet
- Relacion de Personal Observado Por Pase de Lista 2022Document609 pagesRelacion de Personal Observado Por Pase de Lista 2022Alejandro EstudilloNo ratings yet
- 375 e 408 CheetachDocument2 pages375 e 408 CheetachFausto DemicheliNo ratings yet
- Inventario 11-19Document122 pagesInventario 11-19joseNo ratings yet
- SRADocument1 pageSRADee ZeeNo ratings yet
- KEGG PATHWAY - Hsa04216Document4 pagesKEGG PATHWAY - Hsa04216Allie AlieNo ratings yet
- Amount of Water Reqd. For HydrotestDocument1 pageAmount of Water Reqd. For HydrotestRaju VrajuNo ratings yet
- Production Report Summary by EquipmentDocument24 pagesProduction Report Summary by EquipmentAthar KhanNo ratings yet
- B & T Cell (gp63)Document8 pagesB & T Cell (gp63)Jarin Tasnim LiraNo ratings yet
- Exc Kota ProbolinggoDocument20 pagesExc Kota ProbolinggoDea Sarah TiaNo ratings yet
- Lista de Precios Martes 07 - 02 - 2023Document120 pagesLista de Precios Martes 07 - 02 - 2023Jessper SanmiguelNo ratings yet
- ZadacaDocument10 pagesZadacaMuamera HodzicNo ratings yet
- RegressionDocument10 pagesRegressionTilottama SanyalNo ratings yet
- Rituximab Structure - Activity Relationship: Sssequence: Light Chain ChimericDocument4 pagesRituximab Structure - Activity Relationship: Sssequence: Light Chain ChimericSuresh SkNo ratings yet
- R-22 Enthalpy ChartDocument16 pagesR-22 Enthalpy ChartSuhail RanaNo ratings yet
- Genome - A-mtDNA Haplogroup Analysis ReportDocument10 pagesGenome - A-mtDNA Haplogroup Analysis ReportsbzgtNo ratings yet
- Pmwasabi FibrillarinDocument5 pagesPmwasabi FibrillarinAlleleBiotechNo ratings yet
- SUPPLY (1) ImpDocument59 pagesSUPPLY (1) ImpRuchi GuptaNo ratings yet
- pmTFP1 MitochondriaDocument4 pagespmTFP1 MitochondriaAlleleBiotechNo ratings yet
- Flexible BudgetDocument1 pageFlexible BudgetRohan ParimalNo ratings yet
- Batch100 5kDocument85 pagesBatch100 5kEmirTZNo ratings yet
- Latihan StatistikDocument2 pagesLatihan Statistik170254242015 Rahima ZakiaNo ratings yet
- Assembly LineDocument114 pagesAssembly LineFrank RojasNo ratings yet
- SampleDocument18 pagesSampletamilpanniNo ratings yet
- AutosDocument7 pagesAutosRahul BurmanNo ratings yet
- Data Produk 26 01 2024Document22 pagesData Produk 26 01 2024pujiyono83.pyNo ratings yet
- Regression StatisticsDocument21 pagesRegression StatisticsNaimul Haque NayeemNo ratings yet
- Consolidated GSTR - 2A RecoDocument181 pagesConsolidated GSTR - 2A RecokhusbooNo ratings yet
- Img447 CSVDocument15 pagesImg447 CSVNguyen Thi Thuy TienNo ratings yet
- 65 901 ResultsDocument6 pages65 901 ResultsLeyla SaabNo ratings yet
- Zoom CarDocument3 pagesZoom CarSPARROWNo ratings yet
- Dena Bank MIDAS 07032013 To 26032013Document109 pagesDena Bank MIDAS 07032013 To 26032013cratnanamNo ratings yet
- My 23 AndmedtaDocument1,092 pagesMy 23 AndmedtaJoão Pedro Tenório CavalcantiNo ratings yet
- Chamba Luis - 8V0-A - PSKDocument110 pagesChamba Luis - 8V0-A - PSKCarlos SalazarNo ratings yet
- Link Track)Document4 pagesLink Track)Marlon Geronimo100% (1)
- NIDocument2 pagesNIHAMMAMINo ratings yet
- JOHARI Window WorksheetDocument2 pagesJOHARI Window WorksheetAnonymous j9lsM2RBaINo ratings yet
- Emergency War Surgery Nato HandbookDocument384 pagesEmergency War Surgery Nato Handbookboubiyou100% (1)
- BrainPOP Nutrition Quiz242342Document1 pageBrainPOP Nutrition Quiz242342MathableNo ratings yet
- Concrete and Its PropertiesDocument24 pagesConcrete and Its PropertiesAmila LiyanaarachchiNo ratings yet
- CASE 1. Non-Cash Assets Are Sold For P 580,000Document3 pagesCASE 1. Non-Cash Assets Are Sold For P 580,000Riza Mae AlceNo ratings yet
- Cough: A Rapid Expulsion of Air FromDocument2 pagesCough: A Rapid Expulsion of Air FromaubyangNo ratings yet
- Health Module 6 g7Document26 pagesHealth Module 6 g7Richelle PamintuanNo ratings yet
- SpectraSensors TDL Analyzers in RefineriesDocument8 pagesSpectraSensors TDL Analyzers in Refineries1977specopsNo ratings yet
- Ideal Discharge Elderly PatientDocument3 pagesIdeal Discharge Elderly PatientFelicia Risca RyandiniNo ratings yet
- Review Dynamic Earth CoreScienceDocument3 pagesReview Dynamic Earth CoreScienceVikram BologaneshNo ratings yet
- 2-Phase Synchronous-Rectified Buck Controller For Mobile GPU PowerDocument18 pages2-Phase Synchronous-Rectified Buck Controller For Mobile GPU PowerMax Assistência TécnicaNo ratings yet
- Damasco - Cpi - Activity No. 10Document18 pagesDamasco - Cpi - Activity No. 10LDCU - Damasco, Erge Iris M.No ratings yet
- Park Agreement LetterDocument2 pagesPark Agreement LetterKrishna LalNo ratings yet
- Installation and Operation Manual: Proact™ Ii Electric Powered Actuator and DriverDocument32 pagesInstallation and Operation Manual: Proact™ Ii Electric Powered Actuator and DriverDjebali MouradNo ratings yet
- Prestress 3.0Document10 pagesPrestress 3.0Jonel CorbiNo ratings yet
- NFPA 25 2011 Sprinkler Inspection TableDocument2 pagesNFPA 25 2011 Sprinkler Inspection TableHermes VacaNo ratings yet
- Material Specification: Mechanical Property RequirementsDocument2 pagesMaterial Specification: Mechanical Property RequirementsNguyễn Tấn HảiNo ratings yet
- Ethical Consideration in Leadership and ManagementDocument6 pagesEthical Consideration in Leadership and ManagementGlizzle Macaraeg67% (3)
- Aits 2324 Ot I Jeea TD Paper 2 OfflineDocument14 pagesAits 2324 Ot I Jeea TD Paper 2 OfflineAshish SharmaNo ratings yet
- Business Process Dashboard (Raj Mishra)Document22 pagesBusiness Process Dashboard (Raj Mishra)Raj MishraNo ratings yet
- Polyken 4000 PrimerlessDocument2 pagesPolyken 4000 PrimerlessKyaw Kyaw AungNo ratings yet
- Tinh Toan Tang AP Cau Thang - CT Qui LongDocument20 pagesTinh Toan Tang AP Cau Thang - CT Qui Longntt_121987No ratings yet
- Module 5 The Teacher and The Community School Culture and Organizational LeadershipDocument6 pagesModule 5 The Teacher and The Community School Culture and Organizational LeadershipHazeldiazasenas100% (6)
- Pinch & Piston ValvesDocument8 pagesPinch & Piston ValvesJaldhij Patel100% (1)
- Traditional Christmas FoodDocument15 pagesTraditional Christmas FoodAlex DumitracheNo ratings yet
- Inversor Abb 3 8kwDocument2 pagesInversor Abb 3 8kwapi-290643326No ratings yet
- Ems em FW Paneel Firetec enDocument2 pagesEms em FW Paneel Firetec enzlatkokrsicNo ratings yet
- Ryder Quotation 2012.7.25Document21 pagesRyder Quotation 2012.7.25DarrenNo ratings yet
- Pay Structure of Public Employees in PakistanDocument28 pagesPay Structure of Public Employees in PakistanAamir50% (10)
- Ceilcote 222HT Flakeline+ds+engDocument4 pagesCeilcote 222HT Flakeline+ds+englivefreakNo ratings yet
- Gut: the new and revised Sunday Times bestsellerFrom EverandGut: the new and revised Sunday Times bestsellerRating: 4 out of 5 stars4/5 (392)
- Tales from Both Sides of the Brain: A Life in NeuroscienceFrom EverandTales from Both Sides of the Brain: A Life in NeuroscienceRating: 3 out of 5 stars3/5 (18)
- Why We Die: The New Science of Aging and the Quest for ImmortalityFrom EverandWhy We Die: The New Science of Aging and the Quest for ImmortalityRating: 4 out of 5 stars4/5 (3)
- When the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisFrom EverandWhen the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisRating: 3.5 out of 5 stars3.5/5 (2)
- Gut: The Inside Story of Our Body's Most Underrated Organ (Revised Edition)From EverandGut: The Inside Story of Our Body's Most Underrated Organ (Revised Edition)Rating: 4 out of 5 stars4/5 (378)
- Fast Asleep: Improve Brain Function, Lose Weight, Boost Your Mood, Reduce Stress, and Become a Better SleeperFrom EverandFast Asleep: Improve Brain Function, Lose Weight, Boost Your Mood, Reduce Stress, and Become a Better SleeperRating: 4.5 out of 5 stars4.5/5 (15)
- The Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceFrom EverandThe Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceRating: 4.5 out of 5 stars4.5/5 (516)
- A Series of Fortunate Events: Chance and the Making of the Planet, Life, and YouFrom EverandA Series of Fortunate Events: Chance and the Making of the Planet, Life, and YouRating: 4.5 out of 5 stars4.5/5 (62)
- A Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsFrom EverandA Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsRating: 4 out of 5 stars4/5 (5)
- 10% Human: How Your Body's Microbes Hold the Key to Health and HappinessFrom Everand10% Human: How Your Body's Microbes Hold the Key to Health and HappinessRating: 4 out of 5 stars4/5 (33)
- The Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionFrom EverandThe Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionRating: 4 out of 5 stars4/5 (811)
- Undeniable: How Biology Confirms Our Intuition That Life Is DesignedFrom EverandUndeniable: How Biology Confirms Our Intuition That Life Is DesignedRating: 4 out of 5 stars4/5 (11)
- Human: The Science Behind What Makes Your Brain UniqueFrom EverandHuman: The Science Behind What Makes Your Brain UniqueRating: 3.5 out of 5 stars3.5/5 (38)
- All That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesFrom EverandAll That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesRating: 4.5 out of 5 stars4.5/5 (397)
- The Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorFrom EverandThe Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorNo ratings yet
- Who's in Charge?: Free Will and the Science of the BrainFrom EverandWho's in Charge?: Free Will and the Science of the BrainRating: 4 out of 5 stars4/5 (65)
- Good Without God: What a Billion Nonreligious People Do BelieveFrom EverandGood Without God: What a Billion Nonreligious People Do BelieveRating: 4 out of 5 stars4/5 (66)
- Wayfinding: The Science and Mystery of How Humans Navigate the WorldFrom EverandWayfinding: The Science and Mystery of How Humans Navigate the WorldRating: 4.5 out of 5 stars4.5/5 (18)
- Crypt: Life, Death and Disease in the Middle Ages and BeyondFrom EverandCrypt: Life, Death and Disease in the Middle Ages and BeyondRating: 4 out of 5 stars4/5 (4)
- Inside of a Dog: What Dogs See, Smell, and KnowFrom EverandInside of a Dog: What Dogs See, Smell, and KnowRating: 4 out of 5 stars4/5 (390)
- Moral Tribes: Emotion, Reason, and the Gap Between Us and ThemFrom EverandMoral Tribes: Emotion, Reason, and the Gap Between Us and ThemRating: 4.5 out of 5 stars4.5/5 (115)
- The Invention of Tomorrow: A Natural History of ForesightFrom EverandThe Invention of Tomorrow: A Natural History of ForesightRating: 4.5 out of 5 stars4.5/5 (5)
- Buddha's Brain: The Practical Neuroscience of Happiness, Love & WisdomFrom EverandBuddha's Brain: The Practical Neuroscience of Happiness, Love & WisdomRating: 4 out of 5 stars4/5 (215)
- The Second Brain: A Groundbreaking New Understanding of Nervous Disorders of the Stomach and IntestineFrom EverandThe Second Brain: A Groundbreaking New Understanding of Nervous Disorders of the Stomach and IntestineRating: 4 out of 5 stars4/5 (17)