Professional Documents
Culture Documents
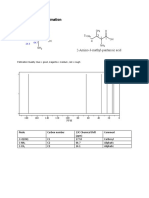
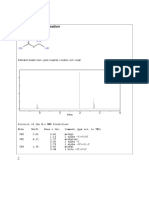
Chemnmr C-13 Estimation: Estimation Quality: Blue Good, Magenta Medium, Red Rough
Uploaded by
firda haqiqiOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Chemnmr C-13 Estimation: Estimation Quality: Blue Good, Magenta Medium, Red Rough
Uploaded by
firda haqiqiCopyright:
Available Formats
ChemNMR C-13 Estimation
32.9
23.1
23.5
134.0
123.4
26.3
H
27.0
Estimation Quality: blue = good, magenta = medium, red = rough
140 120 100 80 60 40 20 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 23.5 -2.3 aliphatic
18.2 2 alpha -C
6.9 1 beta -C=C
9.4 1 beta -C
-2.5 1 gamma -C
-6.2 general corrections
CH2 26.3 -2.3 aliphatic
18.2 2 alpha -C
6.9 1 beta -C=C
9.4 1 beta -C
0.3 1 delta -C
-6.2 general corrections
CH2 27.0 -2.3 aliphatic
19.5 1 alpha -C=C
9.1 1 alpha -C
9.4 1 beta -C
-5.0 2 gamma -C
-3.7 general corrections
CH 123.4 123.3 1-ethylene
17.3 1 -C-C-C-C
-8.9 1 -C-C-C-C
-7.4 1 -C
-0.9 general corrections
C 134.0 123.3 1-ethylene
-8.9 1 -C-C-C-C
17.3 1 -C-C-C-C
9.4 1 -C
-7.1 general corrections
CH2 32.9 -2.3 aliphatic
19.5 1 alpha -C=C
9.1 1 alpha -C
18.8 2 beta -C
-2.5 1 gamma -C
-9.7 general corrections
CH3 23.1 -2.3 aliphatic
19.5 1 alpha -C=C
9.4 1 beta -C
-5.0 2 gamma -C
0.3 1 delta -C
1.2 general corrections
ChemNMR H-1 Estimation
4.75
H
1.96
1.33
H
4.75
1.29 1.96
1.33
Estimation Quality: blue = good, magenta = medium, red = rough
4 3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 1.33 1.37 methylene
-0.04 1 beta -C
0.00 1 beta -C=C
CH2 1.29 1.37 methylene
-0.04 1 beta -C
-0.04 1 beta -C
CH2 1.33 1.37 methylene
-0.04 1 beta -C
0.00 1 beta -C=C
CH2 1.96 1.37 methylene
0.63 1 alpha -C=C
-0.04 1 beta -C
CH2 1.96 1.37 methylene
0.63 1 alpha -C=C
-0.04 1 beta -C
H 4.75 5.25 1-ethylene
-0.50 2 -C c + t
H 4.75 5.25 1-ethylene
-0.50 2 -C c + t
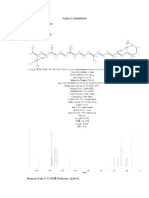
ChemNMR C-13 Estimation
H
39.9
109.1
28.4
H
148.5
27.3 39.9
28.4
Estimation Quality: blue = good, magenta = medium, red = rough
160 140 120 100 80 60 40 20 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 28.4 -2.3 cyclohexane like
18.2 2 alpha -C from aliphatic
6.9 1 beta -C=C from aliphatic
9.4 1 beta -C from aliphatic
-2.5 1 gamma -C from aliphatic
-1.3 general corrections
CH2 27.3 -2.3 cyclohexane like
18.2 2 alpha -C from aliphatic
18.8 2 beta -C from aliphatic
-2.1 1 gamma -C=C from aliphatic
-5.3 general corrections
CH2 28.4 -2.3 cyclohexane like
18.2 2 alpha -C from aliphatic
6.9 1 beta -C=C from aliphatic
9.4 1 beta -C from aliphatic
-2.5 1 gamma -C from aliphatic
-1.3 general corrections
CH2 39.9 -2.3 cyclohexane like
19.5 1 alpha -C=C from aliphatic
9.1 1 alpha -C from aliphatic
18.8 2 beta -C from aliphatic
-2.5 1 gamma -C from aliphatic
-2.7 general corrections
C 148.5 123.3 1-ethylene
34.6 2 -C-C-C-C
-9.4 general corrections
CH2 39.9 -2.3 cyclohexane like
19.5 1 alpha -C=C from aliphatic
9.1 1 alpha -C from aliphatic
18.8 2 beta -C from aliphatic
-2.5 1 gamma -C from aliphatic
-2.7 general corrections
CH2 109.1 123.3 1-ethylene
-17.8 2 -C-C-C-C
3.6 general corrections
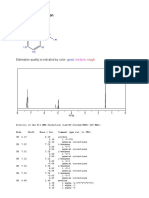
ChemNMR H-1 Estimation
7.19
7.19 7.19
O
O
7.19
5.34 7.19
2.01
Estimation Quality: blue = good, magenta = medium, red = rough
7 6 5 4 3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 7.19 7.26 1-benzene
-0.07 1 -C-O
CH 7.19 7.26 1-benzene
-0.07 1 -C-O
CH 7.19 7.26 1-benzene
-0.07 1 -C-O
CH 7.19 7.26 1-benzene
-0.07 1 -C-O
CH 7.19 7.26 1-benzene
-0.07 1 -C-O
CH2 5.34 1.37 methylene
1.22 1 alpha -1:C*C*C*C*C*C*1
2.75 1 alpha -OC(=O)-C
CH3 2.01 0.86 methyl
1.15 1 alpha -C(=O)OC
ChemNMR C-13 Estimation
129.0
127.2 127.7
O
141.2
O
129.0
170.3
68.2 127.2
20.7
Estimation Quality: blue = good, magenta = medium, red = rough
180 160 140 120 100 80 60 40 20 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
C 141.2 128.5 1-benzene
12.4 1 -C-O
0.3 general corrections
CH 127.2 128.5 1-benzene
-1.2 1 -C-O
-0.1 general corrections
CH 129.0 128.5 1-benzene
0.2 1 -C-O
0.3 general corrections
CH 127.7 128.5 1-benzene
-1.1 1 -C-O
0.3 general corrections
CH 129.0 128.5 1-benzene
0.2 1 -C-O
0.3 general corrections
CH 127.2 128.5 1-benzene
-1.2 1 -C-O
-0.1 general corrections
CH2 68.2 -2.3 aliphatic
24.3 1 alpha -1:C*C*C*C*C*C*1
54.9 1 alpha -O-C=O
-2.5 1 gamma -C
-6.2 general corrections
C 170.3 166.0 1-carboxyl
10.0 1 -C
-5.0 1 -C from O-carboxyl
-0.7 general corrections
CH3 20.7 -2.3 aliphatic
21.8 1 alpha -C(=O)-O
-2.5 1 gamma -C
0.3 1 delta -1:C*C*C*C*C*C*1
3.4 general corrections
ChemNMR H-1 Estimation
3.61 2.0
H OH
2.35
5.60 1.63;1.38
2.35
5.60 1.63;1.38
Estimation Quality: blue = good, magenta = medium, red = rough
5 4 3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 5.60 5.60 cyclopentene
CH 2.35 2.28 cyclopentene
-0.01 1 beta -C from methine
0.08 1 beta -O from methine
CH2 1.63;1.38 1.51 cyclopentane
0.00 1 beta -C=C from methylene
CH 5.60 5.60 cyclopentene
CH 2.35 2.28 cyclopentene
-0.01 1 beta -C from methine
0.08 1 beta -O from methine
CH2 1.63;1.38 1.51 cyclopentane
0.00 1 beta -C=C from methylene
OH 2.0 2.00 alcohol
H 3.61 1.90 cyclopentene
1.73 1 alpha -O from methine
-0.01 1 beta -C from methine
-0.01 1 beta -C from methine
ChemNMR C-13 Estimation
H OH
83.9
50.1
135.9 21.6
50.1
135.9 21.6
Estimation Quality: blue = good, magenta = medium, red = rough
140 120 100 80 60 40 20 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 135.9 135.8 2-norbornene
0.1 general corrections
CH 50.1 -4.5 2-norbornene
19.5 1 alpha -C=C from aliphatic
18.2 2 alpha -C from aliphatic
18.8 2 beta -C from aliphatic
10.1 1 beta -O from aliphatic
-12.0 general corrections
CH2 21.6 -16.6 2-norbornene
18.2 2 alpha -C from aliphatic
6.9 1 beta -C=C from aliphatic
18.8 2 beta -C from aliphatic
-6.2 1 gamma -O from aliphatic
0.5 general corrections
CH 135.9 135.8 2-norbornene
0.1 general corrections
CH 50.1 -4.5 2-norbornene
19.5 1 alpha -C=C from aliphatic
18.2 2 alpha -C from aliphatic
18.8 2 beta -C from aliphatic
10.1 1 beta -O from aliphatic
-12.0 general corrections
CH2 21.6 -16.6 2-norbornene
18.2 2 alpha -C from aliphatic
6.9 1 beta -C=C from aliphatic
18.8 2 beta -C from aliphatic
-6.2 1 gamma -O from aliphatic
0.5 general corrections
CH 83.9 9.9 2-norbornene
18.2 2 alpha -C from aliphatic
49.0 1 alpha -O from aliphatic
6.9 1 beta -C=C from aliphatic
18.8 2 beta -C from aliphatic
-18.9 general corrections
ChemNMR H-1 Estimation
3.38 3.38
Cl Cl
1.85
Estimation Quality: blue = good, magenta = medium, red = rough
3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 3.38 1.37 methylene
2.05 1 alpha -Cl
-0.04 1 beta -C
CH2 1.85 1.37 methylene
0.24 1 beta -Cl
0.24 1 beta -Cl
CH2 3.38 1.37 methylene
2.05 1 alpha -Cl
-0.04 1 beta -C
ChemNMR C-13 Estimation
41.9 41.9
Cl Cl
35.9
Estimation Quality: blue = good, magenta = medium, red = rough
45 40 35 30 25 20 15 10 5 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 41.9 -2.3 aliphatic
9.1 1 alpha -C
31.0 1 alpha -Cl
9.4 1 beta -C
-5.1 1 gamma -Cl
-0.2 general corrections
CH2 35.9 -2.3 aliphatic
18.2 2 alpha -C
20.0 2 beta -Cl
CH2 41.9 -2.3 aliphatic
9.1 1 alpha -C
31.0 1 alpha -Cl
9.4 1 beta -C
-5.1 1 gamma -Cl
-0.2 general corrections
ChemNMR H-1 Estimation
Br
1.74 3.78
1.74
Estimation Quality: blue = good, magenta = medium, red = rough
3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 3.78 1.50 methine
0.34 2 alpha -C
1.94 1 alpha -Br
CH3 1.74 0.86 methyl
0.83 1 beta -Br
0.05 1 beta -C
CH3 1.74 0.86 methyl
0.83 1 beta -Br
0.05 1 beta -C
ChemNMR C-13 Estimation
Br
44.5
29.0
29.0
Estimation Quality: blue = good, magenta = medium, red = rough
45 40 35 30 25 20 15 10 5 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 44.5 -2.3 aliphatic
18.2 2 alpha -C
18.9 1 alpha -Br
9.7 general corrections
CH3 29.0 -2.3 aliphatic
9.1 1 alpha -C
9.4 1 beta -C
11.0 1 beta -Br
1.8 general corrections
CH3 29.0 -2.3 aliphatic
9.1 1 alpha -C
9.4 1 beta -C
11.0 1 beta -Br
1.8 general corrections
ChemNMR H-1 Estimation
2.27 7.07 7.23
O 7.07
1.09
7.07 7.23
Estimation Quality: blue = good, magenta = medium, red = rough
7 6 5 4 3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 7.07 7.26 1-benzene
-0.19 1 -OC(=O)C
CH 7.23 7.26 1-benzene
-0.03 1 -OC(=O)C
CH 7.07 7.26 1-benzene
-0.19 1 -OC(=O)C
CH 7.23 7.26 1-benzene
-0.03 1 -OC(=O)C
CH 7.07 7.26 1-benzene
-0.19 1 -OC(=O)C
CH2 2.27 1.37 methylene
0.00 1 alpha -C
0.90 1 alpha -C(=O)O
CH3 1.09 0.86 methyl
0.23 1 beta -C(=O)O
ChemNMR C-13 Estimation
27.2 172.3 121.6 129.2
O 151.4 125.6
9.4
121.6 129.2
Estimation Quality: blue = good, magenta = medium, red = rough
180 160 140 120 100 80 60 40 20 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
C 151.4 128.5 1-benzene
24.6 1 -O-C(=O)
-1.7 general corrections
CH 121.6 128.5 1-benzene
-7.1 1 -O-C(=O)
0.2 general corrections
CH 129.2 128.5 1-benzene
0.4 1 -O-C(=O)
0.3 general corrections
CH 125.6 128.5 1-benzene
-3.2 1 -O-C(=O)
0.3 general corrections
CH 129.2 128.5 1-benzene
0.4 1 -O-C(=O)
0.3 general corrections
CH 121.6 128.5 1-benzene
-7.1 1 -O-C(=O)
0.2 general corrections
C 172.3 166.0 1-carboxyl
11.0 1 -C-C
-8.0 1 -1:C*C*C*C*C*C*1 from O-carboxyl
3.3 general corrections
CH2 27.2 -2.3 aliphatic
21.8 1 alpha -C(=O)-O
9.1 1 alpha -C
-2.6 1 gamma -1:C*C*C*C*C*C*1
1.2 general corrections
CH3 9.4 -2.3 aliphatic
9.1 1 alpha -C
2.0 1 beta -C(=O)-O
0.3 1 delta -1:C*C*C*C*C*C*1
0.3 general corrections
ChemNMR H-1 Estimation
2.90
H O Cl
H H
4.52
2.65
Estimation Quality: blue = good, magenta = medium, red = rough
4 3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
H 2.90 2.54 oxiran
0.24 1 beta -Cl from methylene
H 2.65 2.54 oxiran
0.24 1 beta -Cl from methylene
H 4.52 2.54 oxiran
1.98 1 alpha -Cl from methine
ChemNMR C-13 Estimation
H O Cl
49.1
64.3
H H
Estimation Quality: blue = good, magenta = medium, red = rough
70 60 50 40 30 20 10 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 49.1 -18.6 oxiran
9.1 1 alpha -C from aliphatic
49.0 1 alpha -O from aliphatic
10.0 1 beta -Cl from aliphatic
-0.4 general corrections
CH 64.3 -18.6 oxiran
9.1 1 alpha -C from aliphatic
31.0 1 alpha -Cl from aliphatic
49.0 1 alpha -O from aliphatic
-6.2 general corrections
ChemNMR H-1 Estimation
2.01
O
1.30
O
4.12
Estimation Quality: blue = good, magenta = medium, red = rough
4 3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH3 1.30 0.86 methyl
0.44 1 beta -OC(=O)C
CH2 4.12 1.37 methylene
0.00 1 alpha -C
2.75 1 alpha -OC(=O)-C
CH3 2.01 0.86 methyl
1.15 1 alpha -C(=O)OC
ChemNMR C-13 Estimation
20.7
O
14.1
170.3
O
61.0
Estimation Quality: blue = good, magenta = medium, red = rough
180 160 140 120 100 80 60 40 20 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH3 14.1 -2.3 aliphatic
9.1 1 alpha -C
6.5 1 beta -O-C=O
0.3 1 delta -C
0.5 general corrections
CH2 61.0 -2.3 aliphatic
9.1 1 alpha -C
54.9 1 alpha -O-C=O
-2.5 1 gamma -C
1.8 general corrections
C 170.3 166.0 1-carboxyl
10.0 1 -C
-5.0 1 -C from O-carboxyl
-0.7 general corrections
CH3 20.7 -2.3 aliphatic
21.8 1 alpha -C(=O)-O
-2.5 1 gamma -C
0.3 1 delta -C
3.4 general corrections
ChemNMR H-1 Estimation
7.21 7.12
3.05
7.08
Br
7.21 7.12 3.63
Estimation Quality: blue = good, magenta = medium, red = rough
7 6 5 4 3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 7.21 7.26 1-benzene
-0.05 1 -CC
CH 7.12 7.26 1-benzene
-0.14 1 -CC
CH 7.12 7.26 1-benzene
-0.14 1 -CC
CH 7.21 7.26 1-benzene
-0.05 1 -CC
CH 7.08 7.26 1-benzene
-0.18 1 -CC
CH2 3.05 1.37 methylene
1.22 1 alpha -1:C*C*C*C*C*C*1
0.46 1 beta -Br
CH2 3.63 1.37 methylene
1.97 1 alpha -Br
0.29 1 beta -1:C*C*C*C*C*C*1
ChemNMR C-13 Estimation
128.7 127.8
139.537.3
126.0
Br
128.7 127.8 31.7
Estimation Quality: blue = good, magenta = medium, red = rough
140 120 100 80 60 40 20 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 128.7 128.5 1-benzene
-0.1 1 -C-C
0.3 general corrections
CH 127.8 128.5 1-benzene
-0.6 1 -C-C
-0.1 general corrections
C 139.5 128.5 1-benzene
11.7 1 -C-C
-0.7 general corrections
CH 127.8 128.5 1-benzene
-0.6 1 -C-C
-0.1 general corrections
CH 128.7 128.5 1-benzene
-0.1 1 -C-C
0.3 general corrections
CH 126.0 128.5 1-benzene
-2.8 1 -C-C
0.3 general corrections
CH2 37.3 -2.3 aliphatic
24.3 1 alpha -1:C*C*C*C*C*C*1
9.1 1 alpha -C
11.0 1 beta -Br
-4.8 general corrections
CH2 31.7 -2.3 aliphatic
9.1 1 alpha -C
18.9 1 alpha -Br
9.3 1 beta -1:C*C*C*C*C*C*1
-3.3 general corrections
ChemNMR H-1 Estimation
O
3.60 3.60
1.60 1.60
1.60
Estimation Quality: blue = good, magenta = medium, red = rough
3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 3.60 3.60 tetrahydropyran
CH2 1.60 1.60 tetrahydropyran
CH2 1.60 1.60 tetrahydropyran
CH2 1.60 1.60 tetrahydropyran
CH2 3.60 3.60 tetrahydropyran
ChemNMR C-13 Estimation
O
69.1 69.1
25.2 25.2
26.0
Estimation Quality: blue = good, magenta = medium, red = rough
70 60 50 40 30 20 10 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 69.1 -4.9 tetrahydropyran
9.1 1 alpha -C from aliphatic
49.0 1 alpha -O from aliphatic
18.8 2 beta -C from aliphatic
-2.5 1 gamma -C from aliphatic
-0.4 general corrections
CH2 25.2 -7.5 tetrahydropyran
18.2 2 alpha -C from aliphatic
9.4 1 beta -C from aliphatic
10.1 1 beta -O from aliphatic
-2.5 1 gamma -C from aliphatic
-2.5 general corrections
CH2 26.0 -5.9 tetrahydropyran
18.2 2 alpha -C from aliphatic
18.8 2 beta -C from aliphatic
-6.2 1 gamma -O from aliphatic
1.1 general corrections
CH2 25.2 -7.5 tetrahydropyran
18.2 2 alpha -C from aliphatic
9.4 1 beta -C from aliphatic
10.1 1 beta -O from aliphatic
-2.5 1 gamma -C from aliphatic
-2.5 general corrections
CH2 69.1 -4.9 tetrahydropyran
9.1 1 alpha -C from aliphatic
49.0 1 alpha -O from aliphatic
18.8 2 beta -C from aliphatic
-2.5 1 gamma -C from aliphatic
-0.4 general corrections
ChemNMR H-1 Estimation
1.00
2.40
2.40
N
1.00
1.00 2.40
Estimation Quality: blue = good, magenta = medium, red = rough
2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 2.40 1.37 methylene
0.00 1 alpha -C
1.03 1 alpha -N(C)C
CH3 1.00 0.86 methyl
0.14 1 beta -N(C)C
CH2 2.40 1.37 methylene
0.00 1 alpha -C
1.03 1 alpha -N(C)C
CH3 1.00 0.86 methyl
0.14 1 beta -N(C)C
CH2 2.40 1.37 methylene
0.00 1 alpha -C
1.03 1 alpha -N(C)C
CH3 1.00 0.86 methyl
0.14 1 beta -N(C)C
ChemNMR C-13 Estimation
13.4
49.2
49.2
N
13.4
13.4 49.2
Estimation Quality: blue = good, magenta = medium, red = rough
50 40 30 20 10 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH2 49.2 -2.3 aliphatic
9.1 1 alpha -C
28.3 1 alpha -N
18.8 2 beta -C
-5.0 2 gamma -C
0.3 general corrections
CH3 13.4 -2.3 aliphatic
9.1 1 alpha -C
11.3 1 beta -N
-5.0 2 gamma -C
0.6 2 delta -C
-0.3 general corrections
CH2 49.2 -2.3 aliphatic
9.1 1 alpha -C
28.3 1 alpha -N
18.8 2 beta -C
-5.0 2 gamma -C
0.3 general corrections
CH3 13.4 -2.3 aliphatic
9.1 1 alpha -C
11.3 1 beta -N
-5.0 2 gamma -C
0.6 2 delta -C
-0.3 general corrections
CH2 49.2 -2.3 aliphatic
9.1 1 alpha -C
28.3 1 alpha -N
18.8 2 beta -C
-5.0 2 gamma -C
0.3 general corrections
CH3 13.4 -2.3 aliphatic
9.1 1 alpha -C
11.3 1 beta -N
-5.0 2 gamma -C
0.6 2 delta -C
-0.3 general corrections
ChemNMR H-1 Estimation
1.71
5.83
H
H
6.88
O 3.76
O
Estimation Quality: blue = good, magenta = medium, red = rough
7 6 5 4 3 2 1 0
PPM
Protocol of the H-1 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
H 5.83 5.25 1-ethylene
-0.22 1 -C cis
0.80 1 -C(=O)O-R gem
CH3 1.71 0.86 methyl
0.85 1 alpha -C=C
H 6.88 5.25 1-ethylene
0.45 1 -C gem
1.18 1 -C(=O)O-R cis
CH3 3.76 0.86 methyl
2.90 1 alpha -OC(=O)-C=C
ChemNMR C-13 Estimation
18.6
124.4 H
142.9
H 166.5
O 52.0
O
Estimation Quality: blue = good, magenta = medium, red = rough
180 160 140 120 100 80 60 40 20 0
PPM
Protocol of the C-13 NMR Prediction:
Node Shift Base + Inc. Comment (ppm rel. to TMS)
CH 142.9 123.3 1-ethylene
9.4 1 -C
5.5 1 -C(=O)OC
4.7 general corrections
CH 124.4 123.3 1-ethylene
-7.4 1 -C
7.1 1 -C(=O)OC
1.4 general corrections
CH3 18.6 -2.3 aliphatic
19.5 1 alpha -C=C
-2.8 1 gamma -C(=O)-O
4.2 general corrections
C 166.5 166.0 1-carboxyl
4.0 1 -C=C-C
-5.0 1 -C from O-carboxyl
1.5 general corrections
CH3 52.0 -2.3 aliphatic
54.9 1 alpha -O-C=O
-2.1 1 gamma -C=C
1.5 general corrections
You might also like
- Chemnmr H-1 Estimation: Nama: Ellen Cristy Ruku Kelas / Nim: PKB 2015 / 15030194100Document26 pagesChemnmr H-1 Estimation: Nama: Ellen Cristy Ruku Kelas / Nim: PKB 2015 / 15030194100vilenNo ratings yet
- Chemnmr C-13 Estimation: 2-Amino-3-Methyl-Pentanoic AcidDocument2 pagesChemnmr C-13 Estimation: 2-Amino-3-Methyl-Pentanoic AcidHanisNo ratings yet
- Acetophenone C-NMR PDFDocument2 pagesAcetophenone C-NMR PDFkobir96No ratings yet
- Tugas 2 ChemdrawDocument14 pagesTugas 2 ChemdrawAshillahldNo ratings yet
- Chemnmr C-13 Estimation: Estimation Quality: Blue Good, Magenta Medium, Red RoughDocument2 pagesChemnmr C-13 Estimation: Estimation Quality: Blue Good, Magenta Medium, Red RoughIcal IrfandyNo ratings yet
- Nama: Skolastika Fera Mulyasari NPM: 11181225 Kelas: 3fa5Document12 pagesNama: Skolastika Fera Mulyasari NPM: 11181225 Kelas: 3fa5galan ryNo ratings yet
- Chemnmr H Estimation: Estimation Quality Is Indicated by ColorDocument2 pagesChemnmr H Estimation: Estimation Quality Is Indicated by ColorHung VuNo ratings yet
- Foto Prosedur PengerjaanDocument29 pagesFoto Prosedur PengerjaanGalan Rizqi YanuarNo ratings yet
- Struktur KimiaDocument4 pagesStruktur KimiaRia Ega LinashitaNo ratings yet
- Chemnmr C Estimation: Estimation Quality Is Indicated by ColorDocument1 pageChemnmr C Estimation: Estimation Quality Is Indicated by ColorStoica AlexandruNo ratings yet
- Senyawa No. 45: RAHMADIANISLAMIATI - A - 08061381722079Document11 pagesSenyawa No. 45: RAHMADIANISLAMIATI - A - 08061381722079Rahma DianNo ratings yet
- A 08061181823019 Rizki PrapitasariDocument53 pagesA 08061181823019 Rizki PrapitasariorinchiaelgaNo ratings yet
- 1-Phenylethanol H-NMR PDFDocument2 pages1-Phenylethanol H-NMR PDFkobir960% (1)
- C C /C R: Qualitative ClarificationDocument3 pagesC C /C R: Qualitative ClarificationAbdelfattah Mohamed OufNo ratings yet
- Acetophenone H NMRDocument2 pagesAcetophenone H NMRkobir96100% (1)
- Assignment 3.A: CH 3143 Reactor EngineeringDocument4 pagesAssignment 3.A: CH 3143 Reactor EngineeringShazni AhamedNo ratings yet
- Jo-Anne P10Document36 pagesJo-Anne P10Jo-Anne St. ClaireNo ratings yet
- CHEE 321: Chemical Reaction Engineering: (Fogler 2.5, 4.3, 8.5) (Fogler 3.2.3, Examples in CH 4 and 8)Document16 pagesCHEE 321: Chemical Reaction Engineering: (Fogler 2.5, 4.3, 8.5) (Fogler 3.2.3, Examples in CH 4 and 8)SchannNo ratings yet
- Alkenes: CC H H H HDocument39 pagesAlkenes: CC H H H Hmanali shahNo ratings yet
- Node 5Document1 pageNode 5HanisNo ratings yet
- CH 09Document32 pagesCH 09Geovanny J.No ratings yet
- Maria Del Mar Bonilla BenavidesDocument5 pagesMaria Del Mar Bonilla Benavidesmaria bonillaNo ratings yet
- Senyawa Ethil Hexyl P-MetoxisinamateDocument9 pagesSenyawa Ethil Hexyl P-Metoxisinamatetika_riskyNo ratings yet
- CBSE Class 12 Chemistry Question Paper Solution 2020 Set 56 1 1Document25 pagesCBSE Class 12 Chemistry Question Paper Solution 2020 Set 56 1 1Saran.kNo ratings yet
- Assignment 1Document11 pagesAssignment 1Hatta AimanNo ratings yet
- A - 08061381823075 - Arrum WardinaDocument68 pagesA - 08061381823075 - Arrum WardinaorinchiaelgaNo ratings yet
- Nama: Arrum Wardina NIM: 08061381823075 Ujian Tengah Semester Review JurnalDocument53 pagesNama: Arrum Wardina NIM: 08061381823075 Ujian Tengah Semester Review JurnalorinchiaelgaNo ratings yet
- A 08061381823101 Bintang Arum LarasatiDocument53 pagesA 08061381823101 Bintang Arum LarasatiorinchiaelgaNo ratings yet
- 06 - Nitrogen Compounds (Level) Module-5Document14 pages06 - Nitrogen Compounds (Level) Module-5Raju SinghNo ratings yet
- 14-Chemistry of HydrocarbonsDocument16 pages14-Chemistry of HydrocarbonsAFTABNo ratings yet
- Sangtaeryang PyoDocument48 pagesSangtaeryang Pyosung dmNo ratings yet
- Chemnmr H-1 Estimation: Estimation Quality: Blue Good, Magenta Medium, Red RoughDocument2 pagesChemnmr H-1 Estimation: Estimation Quality: Blue Good, Magenta Medium, Red RoughSartina Putry SaikhNo ratings yet
- Chem Sample-Midterm-SolutionsDocument12 pagesChem Sample-Midterm-SolutionsScionNo ratings yet
- Tugas Praktikum in Sillico Kmmi: Steven Jeptri Bernandes 21901101095 Class FDocument6 pagesTugas Praktikum in Sillico Kmmi: Steven Jeptri Bernandes 21901101095 Class FStevenNo ratings yet
- MS Chemistry PB XII Set3Document7 pagesMS Chemistry PB XII Set3sahuunnati977No ratings yet
- Reaction Kinetics (Class PDFDocument36 pagesReaction Kinetics (Class PDFUdit GuptaNo ratings yet
- Hydrocarbon SolutionsDocument26 pagesHydrocarbon SolutionskalyanitiffinserviceNo ratings yet
- CBSE Class 12 Chemistry Question Paper Solution 2020 Set 56 1 1Document16 pagesCBSE Class 12 Chemistry Question Paper Solution 2020 Set 56 1 1ram youtubeNo ratings yet
- USP-NF AlcoholDocument4 pagesUSP-NF AlcoholOIL TEST INTERNACIONAL DE COLOMBIA S.A.SNo ratings yet
- Organic Spectroscopy LabDocument17 pagesOrganic Spectroscopy LabcaoimheNo ratings yet
- Chem 51C Worksheet 13Document2 pagesChem 51C Worksheet 13etafeyNo ratings yet
- Chapter 3 Rev1 Rate Laws & StoichiometryDocument35 pagesChapter 3 Rev1 Rate Laws & StoichiometryHakashiMirudoNo ratings yet
- Reactions and Preparations of AlkenesDocument9 pagesReactions and Preparations of AlkenesGolda Meyer VidalNo ratings yet
- Hints and SoluDocument7 pagesHints and Solusedelip276No ratings yet
- New Light Classes: 12B-Che Date-20/01/2022Document2 pagesNew Light Classes: 12B-Che Date-20/01/2022ShashwatNo ratings yet
- Appendix 1: Property Tables and Charts (Si Units)Document50 pagesAppendix 1: Property Tables and Charts (Si Units)Willian KesniNo ratings yet
- CHM207 TutorialDocument3 pagesCHM207 Tutorialit's miaNo ratings yet
- Procedure PlugflowDocument6 pagesProcedure PlugflowhlvijaykumarNo ratings yet
- Exersice PDFDocument24 pagesExersice PDFharsh mishraNo ratings yet
- Alkenes 1Document33 pagesAlkenes 1Abdullah AmjadNo ratings yet
- Combustion Senator. LibyaDocument12 pagesCombustion Senator. Libyasenator.libyaNo ratings yet
- Alkenes Class PresentationDocument68 pagesAlkenes Class PresentationAdityaNo ratings yet
- Chem2exam2 PDFDocument6 pagesChem2exam2 PDFLouis ParrNo ratings yet
- Ban Salclasses: CH MistryDocument7 pagesBan Salclasses: CH MistryApoorv RastogiNo ratings yet
- C-13 NMR InterpretationDocument25 pagesC-13 NMR InterpretationSudipta SahaNo ratings yet
- Alkyl Halides and Amines Mcqs KeyDocument3 pagesAlkyl Halides and Amines Mcqs KeySameer HussainNo ratings yet
- CM2121 Tutorial 4 (AY 21-22 Sem 1)Document4 pagesCM2121 Tutorial 4 (AY 21-22 Sem 1)barry allenNo ratings yet
- Homework 4 QuestionDocument1 pageHomework 4 QuestionTouqeer iqbalNo ratings yet
- (Extra Copy) Lecture 2Document31 pages(Extra Copy) Lecture 2Udit GuptaNo ratings yet
- Allied Chemistry 2017-18-27082018Document6 pagesAllied Chemistry 2017-18-27082018Masum BillahNo ratings yet
- AY2324 - Teachers - Administration GR 11Document16 pagesAY2324 - Teachers - Administration GR 11krisnuNo ratings yet
- Preparation of Acyl HalidesDocument34 pagesPreparation of Acyl HalidesOktavia Eka ArdyantiNo ratings yet
- Syn, Anti, or Random and The Stereochemistry of The Starting Alkene, Various Stereoisomeric Products WillDocument6 pagesSyn, Anti, or Random and The Stereochemistry of The Starting Alkene, Various Stereoisomeric Products WillLucy grahamNo ratings yet
- Evaporation Rate of Solvents PDFDocument2 pagesEvaporation Rate of Solvents PDFsyamlokNo ratings yet
- Comprehensive Cellulose Chemistry - Vol. 2 - Functionalization of CelluloseDocument417 pagesComprehensive Cellulose Chemistry - Vol. 2 - Functionalization of CelluloserodsNo ratings yet
- Angew. Chem. AzulenesDocument4 pagesAngew. Chem. AzulenesveromendoNo ratings yet
- Alkaloids: DR N AhmedDocument23 pagesAlkaloids: DR N AhmedMohammad SamirNo ratings yet
- Alkenes, Alkynes, and Addition ReactionsDocument23 pagesAlkenes, Alkynes, and Addition Reactionsapi-37062900% (1)
- Name: Pranav Arun Patil Roll No: A007Document20 pagesName: Pranav Arun Patil Roll No: A007Pranav PatilNo ratings yet
- Technical Note TN-106: Correction Factors, Ionization Energies, and Calibration CharacteristicsDocument12 pagesTechnical Note TN-106: Correction Factors, Ionization Energies, and Calibration Characteristicssrinivas1108No ratings yet
- CBSE Class 12 Chemistry - Amines Chapter NotesDocument12 pagesCBSE Class 12 Chemistry - Amines Chapter NotesGayathiriNo ratings yet
- 3B-HYDROCARBON Assignment - FinalDocument49 pages3B-HYDROCARBON Assignment - Finalkraken monsterNo ratings yet
- B.SC I Organo Sulphur CompoundsDocument3 pagesB.SC I Organo Sulphur CompoundsNickOoPandeyNo ratings yet
- Amines AmidesDocument17 pagesAmines AmidesApril EnoslayNo ratings yet
- Preparation of Benzoic Acid Using A Grignard Reagent: Experiment 5Document9 pagesPreparation of Benzoic Acid Using A Grignard Reagent: Experiment 5mamadou17diallo17No ratings yet
- CHM243 Final Exam #2 2011Document15 pagesCHM243 Final Exam #2 2011XiaohanJinNo ratings yet
- Organic Chemistry: Exercise - I Exercise - Ii Exercise-Iii Exercise Iv Answer KeyDocument39 pagesOrganic Chemistry: Exercise - I Exercise - Ii Exercise-Iii Exercise Iv Answer KeyRaju SinghNo ratings yet
- A Primer To Designing Organic SynthesisDocument42 pagesA Primer To Designing Organic SynthesisMohammed100% (1)
- Synthesis of Schiff Bases by Organic Free Solvent MethodDocument4 pagesSynthesis of Schiff Bases by Organic Free Solvent Methodlox agencyNo ratings yet
- Carbon and Its CompoundsDocument22 pagesCarbon and Its Compoundsapi-24735088294% (17)
- General Organic ChemistryDocument29 pagesGeneral Organic ChemistryBombay TechnologyNo ratings yet
- Physical Science Q1 Module 4Document22 pagesPhysical Science Q1 Module 4Alfred RodellasNo ratings yet
- Syllabus Diploma (Cse)Document79 pagesSyllabus Diploma (Cse)Dhirendra YadavNo ratings yet
- 12th Class Chemistry Chapter 8Document49 pages12th Class Chemistry Chapter 8ailaNo ratings yet
- Organic Chem U-3 and 4Document77 pagesOrganic Chem U-3 and 4sinte beyuNo ratings yet
- Intermediate Mediocon Product ListDocument43 pagesIntermediate Mediocon Product ListAnand SolomonNo ratings yet
- Synthesis of Dothiepin and Doxepin by Grignard Reactions in TolueneDocument6 pagesSynthesis of Dothiepin and Doxepin by Grignard Reactions in TolueneJuan Carlos VillotaNo ratings yet
- Experiment 8 Hydrocarbons: CHY52 (Organic Chemistry I) MF (1:00 PM - 4:00 PM)Document13 pagesExperiment 8 Hydrocarbons: CHY52 (Organic Chemistry I) MF (1:00 PM - 4:00 PM)jestoni langgido100% (1)
- Chem 243 Midterm ReviewDocument152 pagesChem 243 Midterm ReviewProfAnonymous100% (1)