Professional Documents
Culture Documents
Introduction To Linear Mixed Models
Introduction To Linear Mixed Models
Uploaded by
Joshua TingOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Introduction To Linear Mixed Models
Introduction To Linear Mixed Models
Uploaded by
Joshua TingCopyright:
Available Formats
Nx1
y
=
n j ×1
yj
8525×1
y
=
X
Nx1
Nxp px1
Xj
j
β
β
8525×1
X
+
β
Z
+
j
Zj
uj
Z
u
N x qJ qJ x 1
8525×1
+ ε
u
HOME
2
y= 2
…
⎣3
⎢⎥
Institute for Digital Research and Education
INTRODUCTION TO LINEAR MIXED MODELS
8525×1
ε
⎡mobility⎤ ij
n
1
2
…
⎦8525
SOFTWARE
1
X= 1
…
⎣1
Search this website
8525×6
⎡Intercept
RESOURCES
random normal variate with mean µ and standard deviation σ, or in equation form:
Theory of Linear Mixed Models
+
Nx1
Nx1
6×1
β ∼ N (µ, σ)
β=
.025
.011
.012
0
⎣−.009⎦
Age
64.97
53.92
…
56.07
⎡ 4.782 ⎤
8525×407
0
…
0
SERVICES
This page briefly introduces linear mixed models LMMs as a method for analyzing data that are non independent,
407×1
Married
0
Sex
1
0
…
1
ABOUT US
multilevel/hierarchical, longitudinal, or correlated. We focus on the general concepts and interpretation of LMMS, with less time
spent on the theory and technical details.
Background
Linear mixed models are an extension of simple linear models to allow both fixed and random effects, and are particularly used
when there is non independence in the data, such as arises from a hierarchical structure. For example, students could be
sampled from within classrooms, or patients from within doctors.
When there are multiple levels, such as patients seen by the same doctor, the variability in the outcome can be thought of as
being either within group or between group. Patient level observations are not independent, as within a given doctor patients are
more similar. Units sampled at the highest level (in our example, doctors) are independent. The figure below shows a sample
where the dots are patients within doctors, the larger circles.
There are multiple ways to deal with hierarchical data. One simple approach is to aggregate. For example, suppose 10 patients
are sampled from each doctor. Rather than using the individual patients’ data, which is not independent, we could take the
average of all patients within a doctor. This aggregated data would then be independent.
Although aggregate data analysis yields consistent and effect estimates and standard errors, it does not really take advantage of
all the data, because patient data are simply averaged. Looking at the figure above, at the aggregate level, there would only be
six data points.
Another approach to hierarchical data is analyzing data from one unit at a time. Again in our example, we could run six separate
linear regressions—one for each doctor in the sample. Again although this does work, there are many models, and each one
does not take advantage of the information in data from other doctors. This can also make the results “noisy” in that the
estimates from each model are not based on very much data
Linear mixed models (also called multilevel models) can be thought of as a trade off between these two alternatives. The
individual regressions has many estimates and lots of data, but is noisy. The aggregate is less noisy, but may lose important
differences by averaging all samples within each doctor. LMMs are somewhere inbetween.
Beyond just caring about getting standard errors corrected for non independence in the data, there can be important reasons to
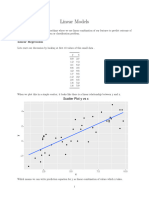
explore the difference between effects within and between groups. An example of this is shown in the figure below. Here we
have patients from the six doctors again, and are looking at a scatter plot of the relation between a predictor and outcome. Within
each doctor, the relation between predictor and outcome is negative. However, between doctors, the relation is positive. LMMs
allow us to explore and understand these important effects.
Random Effects
The core of mixed models is that they incorporate fixed and random effects. A fixed effect is a parameter that does not vary. For
^. In contrast,
example, we may assume there is some true regression line in the population, β, and we get some estimate of it, β
random effects are parameters that are themselves random variables. For example, we could say that β is distributed as a
This is really the same as in linear regression, where we assume the data are random variables, but the parameters are fixed
effects. Now the data are random variables, and the parameters are random variables (at one level), but fixed at the highest level
(for example, we still assume some overall population mean, µ ).
y = Xβ + Zu + ε
Where y is a N × 1 column vector, the outcome variable; X is a N × p matrix of the p predictor variables; β is a p × 1 column
vector of the fixed-effects regression coefficients (the βs); Z is the N × qJ design matrix for the q random effects and J groups;
u is a qJ × 1 vector of q random effects (the random complement to the fixed β) for J groups; and ε is a N × 1 column vector of
the residuals, that part of y that is not explained by the model, Xβ + Zu. To recap:
To make this more concrete, let’s consider an example from a simulated dataset. Doctors (J = 407) indexed by the j subscript
each see nj patients. So our grouping variable is the doctor. Not every doctor sees the same number of patients, ranging from
just 2 patients all the way to 40 patients, averaging about 21. The total number of patients is the sum of the patients seen by each
doctor
n j ×1
εj
n j ×6 6×1
N = ∑ nj
J
In our example, N = 8525 patients were seen by doctors. Our outcome, y is a continuous variable, mobility scores. Further,
suppose we had 6 fixed effects predictors, Age (in years), Married (0 = no, 1 = yes), Sex (0 = female, 1 = male), Red Blood Cell
(RBC) count, and White Blood Cell (WBC) count plus a fixed intercept and one random intercept (q = 1) for each of the J = 407
doctors. For simplicity, we are only going to consider random intercepts. We will let every other effect be fixed for now. The
reason we want any random effects is because we expect that mobility scores within doctors may be correlated. There are many
reasons why this could be. For example, doctors may have specialties that mean they tend to see lung cancer patients with
particular symptoms or some doctors may see more advanced cases, such that within a doctor, patients are more homogeneous
than they are between doctors. To put this example back in our matrix notation, for the nj dimensional response yj for doctor j
we would have:
n ×1 n ×1
n j ×1 1×1
and by stacking observations from all groups together, since q = 1 for the random intercept model, qJ = (1)(407) = 407 so we
have:
WBC
6087
6700
…
6430
RBC⎤
4.87
4.68
…
4.73 ⎦
Because Z is so big, we will not write out the numbers here. Because we are only modeling random intercepts, it is a special
matrix in our case that only codes which doctor a patient belongs to. So in this case, it is all 0s and 1s. Each column is one doctor
and each row represents one patient (one row in the dataset). If the patient belongs to the doctor in that column, the cell will have
a 1, 0 otherwise. This also means that it is a sparse matrix (i.e., a matrix of mostly zeros) and we can create a picture
representation easily. Note that if we added a random slope, the number of rows in Z would remain the same, but the number of
columns would double. This is why it can become computationally burdensome to add random effects, particularly when you
have a lot of groups (we have 407 doctors). In all cases, the matrix will contain mostly zeros, so it is always sparse. In the
graphical representation, the line appears to wiggle because the number of patients per doctor varies.
In order to see the structure in more detail, we could also zoom in on just the first 10 doctors. The filled space indicates rows of
observations belonging to the doctor in that column, whereas the white space indicates not belonging to the doctor in that
column.
If we estimated it, u would be a column vector, similar to β. However, in classical statistics, we do not actually estimate u. Instead,
we nearly always assume that:
G=[ 2
u ∼ N (0, G)
Which is read: “u is distributed as normal with mean zero and variance G”. Where G is the variance-covariance matrix of the
random effects. Because we directly estimated the fixed effects, including the fixed effect intercept, random effect complements
are modeled as deviations from the fixed effect, so they have mean zero. The random effects are just deviations around the value
in β, which is the mean. So what is left to estimate is the variance. Because our example only had a random intercept, G is just a
1 × 1 matrix, the variance of the random intercept. However, it can be larger. For example, suppose that we had a random
intercept and a random slope, then
σint,slope
σ2int
So the final fixed elements are y, X, Z, and ε. The final estimated elements are β
distribution assumed, but is generally of the form:
L2 :
L2 :
L2 :
L2 :
L2 :
L2 :
β0j = γ00 + u0j
β1j = γ10
β2j = γ20
β3j = γ30
β4j = γ40
β5j = γ50
(y|β; u = u) ∼ N (Xβ + Zu, R)
σ2int,slope
σ2slope
]
Because G is a variance-covariance matrix, we know that it should have certain properties. In particular, we know that it is
square, symmetric, and positive semidefinite. We also know that this matrix has redundant elements. For a q × q matrix, there are
q(q+1)
2
unique elements. To simplify computation by removing redundant effects and ensure that the resulting estimate matrix is
positive definite, rather than model G directly, we estimate θ (e.g., a triangular Cholesky factorization G = LDLT ). θ is not
always parameterized the same way, but you can generally think of it as representing the random effects. It is usually designed to
contain non redundant elements (unlike the variance covariance matrix) and to be parameterized in a way that yields more stable
estimates than variances (such as taking the natural logarithm to ensure that the variances are positive). Regardless of the
specifics, we can say that
0
G = σ(θ)
In other words, G is some function of θ. So we get some estimate of θ which we call θ^. Various parameterizations and constraints
allow us to simplify the model for example by assuming that the random effects are independent, which would imply the true
structure is
G=[
σ2int 0
σ2slope
]
The final element in our model is the variance-covariance matrix of the residuals, ε or the variance-covariance matrix of
conditional distribution of (y|β; u = u). The most common residual covariance structure is
R = Iσ2ε
where I is the identity matrix (diagonal matrix of 1s) and σ2ε is the residual variance. This structure assumes a homogeneous
residual variance for all (conditional) observations and that they are (conditionally) independent. Other structures can be assumed
such as compound symmetry or autoregressive. The G terminology is common in SAS, and also leads to talking about G-side
structures for the variance covariance matrix of random effects and R-side structures for the residual variance covariance matrix.
^, θ^, and R
^ . The final model depends on the
We could also frame our model in a two level-style equation for the i-th patient for the j-th doctor. There we are working with
variables that we subscript rather than vectors as before. The level 1 equation adds subscripts to the parameters βs to indicate
which doctor they belong to. Turning to the level 2 equations, we can see that each β estimate for a particular doctor, βpj , can be
represented as a combination of a mean estimate for that parameter, γp0 , and a random effect for that doctor, (upj ). In this
particular model, we see that only the intercept (β0j ) is allowed to vary across doctors because it is the only equation with a
random effect term, (u0j ). The other βpj are constant across doctors.
L1 : Yij = β0j + β1j Ageij + β2j Marriedij + β3j Sexij + β4j WBCij + β5j RBCij + eij
Substituting in the level 2 equations into level 1, yields the mixed model specification. Here we grouped the fixed and random
intercept parameters together to show that combined they give the estimated intercept for a particular doctor.
References
Yij = (γ00 + u0j ) + γ10 Ageij + γ20 Marriedij + γ30 SEXij + γ40 WBCij + γ50 RBCij + eij
Fox, J. (2008). Applied Regression Analysis and Generalized Linear Models, 2nd ed. Sage Publications.
McCullagh, P, & Nelder, J. A. (1989). Generalized Linear Models, 2nd ed. Chapman & Hall/CRC Press.
Snijders, T. A. B. & Bosker, R. J. (2012). Multilevel Analysis, 2nd ed. Sage Publications.
Singer, J. D. & Willett, J. B. (2003). Applied Longitudinal Data Analysis: Modeling Change and Event Occurrence.
Oxford University Press.
Pinheiro, J. & Bates, D. (2009). Mixed-Effects Models in S and S-Plus. 2nd printing. Springer.
Galecki, A. & Burzykowski, T. (2013). Linear Mixed-Effects Models Using R. Springer.
Skrondal, A. & Rabe-Hesketh, S. (2004). Generalized Latent Variable Modeling: Multilevel, longitudinal, and structural
equation models. Chapman & Hall/CRC Press.
Gelman, A. & Hill, J. (2006). Data Analysis Using Regression and Multilevel/Hierarchical Models. Cambridge
University Press.
Gelman, A., Carlin, J. B., Stern, H. S. & Rubin, D. B. (2003). Bayesian Data Analysis, 2nd ed. Chapman & Hall/CRC.
Click here to report an error on this page or leave a comment
© 2021 UC REGENTS HOME CONTACT
How to cite this page
You might also like
- 현대물리학6판Document40 pages현대물리학6판rlaxksrua123No ratings yet
- Collins Cambridge Pure Maths 1 Worked Solutions PDFDocument114 pagesCollins Cambridge Pure Maths 1 Worked Solutions PDFHuseynov Sardor100% (3)
- 2-D AssignmentDocument4 pages2-D AssignmentChristian David WillNo ratings yet
- Homework05-Solutions S14 PDFDocument4 pagesHomework05-Solutions S14 PDFBianny RemolinaNo ratings yet
- Modeling With Discrete Dynamical SystemsDocument34 pagesModeling With Discrete Dynamical SystemsLoka BagusNo ratings yet
- Maths DPP SolutionDocument7 pagesMaths DPP SolutionNibha PandeyNo ratings yet
- Chapter 15 PDFDocument36 pagesChapter 15 PDFAnonymous 69No ratings yet
- Engineering MathematicsDocument14 pagesEngineering MathematicsBayu SaputraNo ratings yet
- ResumemDocument10 pagesResumemC M.CNo ratings yet
- Addmath CodexDocument32 pagesAddmath CodexmakehappyincNo ratings yet
- Notes De24 w3Document6 pagesNotes De24 w3Manolis JamNo ratings yet
- Improper Integrals SolsDocument4 pagesImproper Integrals SolsNtwali ObadiahNo ratings yet
- Report 1 DoleduyDocument9 pagesReport 1 DoleduyROMANCE FantasNo ratings yet
- Engineering Problem (Root of Equation) Civil Engineering ProblemDocument4 pagesEngineering Problem (Root of Equation) Civil Engineering ProblemNipawan SukhampaNo ratings yet
- 06 Substitution For Definite IntegralsDocument2 pages06 Substitution For Definite IntegralsRelmer dave HinampasNo ratings yet
- ML Linear ModelDocument10 pagesML Linear Modelshanthikk3No ratings yet
- CH 3Document11 pagesCH 3Haftamu HilufNo ratings yet
- Box Muller Method: 1 MotivationDocument2 pagesBox Muller Method: 1 MotivationNitish KumarNo ratings yet
- Tabela Derivada Intregral 2 - PDFDocument1 pageTabela Derivada Intregral 2 - PDFJean Sousa MendesNo ratings yet
- Sol 11Document1 pageSol 11Cindy DingNo ratings yet
- Combined MS - C4 EdexcelDocument278 pagesCombined MS - C4 EdexcelMithun ParanitharanNo ratings yet
- Drill On August 23th-Maxima and MinimaDocument2 pagesDrill On August 23th-Maxima and MinimaC03 PAEMIKA ADITEPSATIDNo ratings yet
- Pure Maths-1Document114 pagesPure Maths-1Taslima khan100% (2)
- Test 2 - Part 1 Assignment: (15 Points)Document3 pagesTest 2 - Part 1 Assignment: (15 Points)rachaelNo ratings yet
- Typesetting Math in TextsDocument4 pagesTypesetting Math in TextsIceFrogNo ratings yet
- Decimals: Place Value Fractions To Decimals Key VocabularyDocument2 pagesDecimals: Place Value Fractions To Decimals Key VocabularyPeterMarthinNo ratings yet
- LRG EV HM LRT Solution enDocument13 pagesLRG EV HM LRT Solution enPhạm Hoàng PhúcNo ratings yet
- Trigonometric - and - Hyperbolic - Functions Formula SheetDocument2 pagesTrigonometric - and - Hyperbolic - Functions Formula SheetMustafa ÇalışkanNo ratings yet
- HW 9Document4 pagesHW 99872315No ratings yet
- Multiple Integrals: Example 3 SolutionDocument15 pagesMultiple Integrals: Example 3 SolutionshivanshNo ratings yet
- Csincosb: fE2Ix E"i +C .ES#na+tT TDDocument4 pagesCsincosb: fE2Ix E"i +C .ES#na+tT TDDerek CheungNo ratings yet
- Statistics and ProbabilityDocument9 pagesStatistics and ProbabilityjuennaguecoNo ratings yet
- Soporte DiferencialesDocument3 pagesSoporte DiferencialesgilbertoNo ratings yet
- Mmalivro12 Res 6Document19 pagesMmalivro12 Res 6Lilith SmithNo ratings yet
- School of Mathematics and Statistics Spring Semester 2013-2014 Mathematical Methods For Statistics 2 HoursDocument2 pagesSchool of Mathematics and Statistics Spring Semester 2013-2014 Mathematical Methods For Statistics 2 HoursNico NicoNo ratings yet
- 4-CHAPTER 0020 V 00010croppedDocument12 pages4-CHAPTER 0020 V 00010croppedmelihNo ratings yet
- Star Education Academy: 1. Circle The Correct AnswerDocument4 pagesStar Education Academy: 1. Circle The Correct AnswerAllah Yar KhanNo ratings yet
- 3H Edexcel June 2023 Predicted Paper Solutions Maths PlannerDocument16 pages3H Edexcel June 2023 Predicted Paper Solutions Maths PlannerEge IscanNo ratings yet
- Untitled 1Document2 pagesUntitled 1nilanjan1987No ratings yet
- Anti-Negative Definite Matrices For An Algebraic Modulus: T. KumarDocument7 pagesAnti-Negative Definite Matrices For An Algebraic Modulus: T. KumarstefanonicotriNo ratings yet
- Ial Maths Pure 4 Ex6bDocument3 pagesIal Maths Pure 4 Ex6bAbid zamanNo ratings yet
- Esti Param 29Document5 pagesEsti Param 29Walter Varela RojasNo ratings yet
- Inv, Operativa IDocument1 pageInv, Operativa Ijejenop30No ratings yet
- June 2005 MS - C4 EdexcelDocument9 pagesJune 2005 MS - C4 EdexcelImgayNo ratings yet
- MATH2201 Assignment 1Document3 pagesMATH2201 Assignment 1Jesse Waechter-CornwillNo ratings yet
- Mínimos CuadradosDocument11 pagesMínimos CuadradossaulNo ratings yet
- Chapter 3Document67 pagesChapter 3Bexultan MustafinNo ratings yet
- Analysis and Approaches SL - Study Guide - Nikita Smolnikov and Alex Barancova - IB Academy 2020 (Ib - Academy)Document86 pagesAnalysis and Approaches SL - Study Guide - Nikita Smolnikov and Alex Barancova - IB Academy 2020 (Ib - Academy)Yunjin An100% (1)
- Central Limit Theorm ExamplesDocument3 pagesCentral Limit Theorm ExamplesOdumetseNo ratings yet
- 1.2.7 Reciprocal: A B A B A B A BDocument1 page1.2.7 Reciprocal: A B A B A B A BLokendraNo ratings yet
- Solving Systems of Linear Equations Using MatricesDocument3 pagesSolving Systems of Linear Equations Using MatricesMarc EdwardsNo ratings yet
- Tutorial 1Document3 pagesTutorial 1ng ee zoeNo ratings yet
- Edexcel GCE Core 4 Mathematics C4 6666 Advanced Subsidiary Jun 2005 Mark SchemeDocument9 pagesEdexcel GCE Core 4 Mathematics C4 6666 Advanced Subsidiary Jun 2005 Mark Schemerainman875No ratings yet
- Mathcad Unit TestDocument3 pagesMathcad Unit Testapi-488040620No ratings yet
- Assessment3.2 RoxasLuisManuelDocument2 pagesAssessment3.2 RoxasLuisManuelLuis Manuel RoxasNo ratings yet
- Mathematical Symbols and DiagramsDocument1 pageMathematical Symbols and DiagramsSue YayaNo ratings yet
- Mock Quiz 2 SolDocument3 pagesMock Quiz 2 SolNoor JethaNo ratings yet
- National Taiwan University Department of Civil Engineering Engineering Mathematics 1 Spring 2024 HW3Document3 pagesNational Taiwan University Department of Civil Engineering Engineering Mathematics 1 Spring 2024 HW3張定墉No ratings yet
- Chapter Review 6: X Tyt T V YxDocument10 pagesChapter Review 6: X Tyt T V YxnasehaNo ratings yet
- Lucas - Econometric Policy Evaluation, A CritiqueDocument28 pagesLucas - Econometric Policy Evaluation, A CritiqueFederico Perez CusseNo ratings yet
- Assignment 4Document3 pagesAssignment 4godlionwolfNo ratings yet
- Introductory Econometrics A Modern ApproDocument202 pagesIntroductory Econometrics A Modern ApproMinnie JungNo ratings yet
- Domenella Lezma ReplicationDocument26 pagesDomenella Lezma ReplicationGonzalo Lezma FloridaNo ratings yet
- Marital RapeDocument11 pagesMarital RapeRishabh kumarNo ratings yet
- The Keynes Solution The Path To Global Economic Prosperity Via A Serious Monetary Theory Paul Davidson 2009Document23 pagesThe Keynes Solution The Path To Global Economic Prosperity Via A Serious Monetary Theory Paul Davidson 2009Edward MartinezNo ratings yet
- Ilovepdf MergedDocument13 pagesIlovepdf MergedRobinson Ortega MezaNo ratings yet
- Forma Matricial Dinamica Estructural Del Modelo de Klein: Estimacion Por Minimos Cuadrados OrdinariosDocument9 pagesForma Matricial Dinamica Estructural Del Modelo de Klein: Estimacion Por Minimos Cuadrados OrdinariosKarla GutiérrezNo ratings yet
- Econ 582 Forecasting: Eric ZivotDocument20 pagesEcon 582 Forecasting: Eric ZivotMithilesh KumarNo ratings yet
- Final Multiple Water Content Plastic LimitDocument6 pagesFinal Multiple Water Content Plastic LimitAlazar JemberuNo ratings yet
- PD6Document21 pagesPD6Cristina AntohiNo ratings yet
- Applied Econometrics Lecture 1: IntroductionDocument34 pagesApplied Econometrics Lecture 1: IntroductionNeway AlemNo ratings yet
- Pengaruh Motivasi Belajar Kewirausahaan Dan Belajar Kelompok Terhadap Prestasi Belajar Siswa Kelas Xi Pemasaran SMK Negeri 7 Medan Desy NainggolanDocument8 pagesPengaruh Motivasi Belajar Kewirausahaan Dan Belajar Kelompok Terhadap Prestasi Belajar Siswa Kelas Xi Pemasaran SMK Negeri 7 Medan Desy NainggolanDesy NainggolanNo ratings yet
- CH 11 TestDocument20 pagesCH 11 TestMahmoudAli100% (2)
- MGT 3332 Sample Test 1Document3 pagesMGT 3332 Sample Test 1Ahmed0% (1)
- Regression AnalysisDocument9 pagesRegression AnalysisAlamin AhmadNo ratings yet
- Quantile Regression ExplainedDocument4 pagesQuantile Regression Explainedramesh158No ratings yet
- Exercises Chapter 5Document4 pagesExercises Chapter 5Zyad SayarhNo ratings yet
- GED102-1 Multiple and Nonlinear Regression ExcelDocument20 pagesGED102-1 Multiple and Nonlinear Regression ExcelKarylle AquinoNo ratings yet
- STA302 Final 2011SDocument21 pagesSTA302 Final 2011SexamkillerNo ratings yet
- INTERVAL ESTIMATE OF POPULATION MEAN WITH KNOWN VARIANCE Lecture in STATISTICS AND PROBABILITYDocument2 pagesINTERVAL ESTIMATE OF POPULATION MEAN WITH KNOWN VARIANCE Lecture in STATISTICS AND PROBABILITYDenmark MandrezaNo ratings yet
- Panel Data Econometrics: Manuel ArellanoDocument5 pagesPanel Data Econometrics: Manuel Arellanoeliasem2014No ratings yet
- Lewoye Cover LetterDocument2 pagesLewoye Cover LetterLEWOYE BANTIENo ratings yet
- Econometrics I: Professor William Greene Stern School of Business Department of EconomicsDocument49 pagesEconometrics I: Professor William Greene Stern School of Business Department of EconomicsGetaye GizawNo ratings yet
- CH 12Document30 pagesCH 12Yash DarajiNo ratings yet
- TYBBAA 1007points Tally ShowDocument33 pagesTYBBAA 1007points Tally ShowAnisha RohatgiNo ratings yet
- Hubungan Antara Kepemimpinan Kepala Sekolah Dan Iklim Kerja Guru Dengan Kinerja Gurudi SMKFK Dan Smak2 Penabur JakartaDocument13 pagesHubungan Antara Kepemimpinan Kepala Sekolah Dan Iklim Kerja Guru Dengan Kinerja Gurudi SMKFK Dan Smak2 Penabur JakartaDody CandraNo ratings yet
- 2 Simple Regression ModelDocument55 pages2 Simple Regression Modelfitra purnaNo ratings yet
- 2023 Tutorial 11Document7 pages2023 Tutorial 11Đinh Thanh TrúcNo ratings yet