Professional Documents
Culture Documents
Homology Modeling: Ref: Structural Bioinformatics, P.E Bourne Molecular Modeling, Folkers
Uploaded by
Pallavi Verma0 ratings0% found this document useful (0 votes)
40 views16 pagesThe structure of a protein is uniquely determined by its amino acid sequence. The structure is more stable and changes much slower than the associated sequence. Significant differences in protein structures occur preferably in loop regions, structurally variable regions. Loop search method, looks for peptide segments which meet the specified geometrical criterion.
Original Description:
Original Title
Homology Model
Copyright
© Attribution Non-Commercial (BY-NC)
Available Formats
PPT, PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentThe structure of a protein is uniquely determined by its amino acid sequence. The structure is more stable and changes much slower than the associated sequence. Significant differences in protein structures occur preferably in loop regions, structurally variable regions. Loop search method, looks for peptide segments which meet the specified geometrical criterion.
Copyright:
Attribution Non-Commercial (BY-NC)
Available Formats
Download as PPT, PDF, TXT or read online from Scribd
0 ratings0% found this document useful (0 votes)
40 views16 pagesHomology Modeling: Ref: Structural Bioinformatics, P.E Bourne Molecular Modeling, Folkers
Uploaded by
Pallavi VermaThe structure of a protein is uniquely determined by its amino acid sequence. The structure is more stable and changes much slower than the associated sequence. Significant differences in protein structures occur preferably in loop regions, structurally variable regions. Loop search method, looks for peptide segments which meet the specified geometrical criterion.
Copyright:
Attribution Non-Commercial (BY-NC)
Available Formats
Download as PPT, PDF, TXT or read online from Scribd
You are on page 1of 16
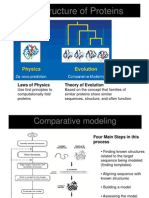
Homology Modeling
Ref: Structural bioinformatics, P.E Bourne
Molecular modeling, Folkers
“To predict a structure from its sequence with
an accuracy that is comparable to the best
results achieved experimentally.”
It is based on two major observations:
1. The structure of a protein is uniquely
determined by its amino acid sequence.
Knowing the sequence should suffice to
obtain the structure.
2. During evolution, the structure is more stable
and changes much slower than the associated
sequence, so that similar sequences adopt
practically identical structures, and distantly
related sequences still fold into similar
structures.
• 1. Template recognition and initial alignment
• 2. Alignment correction
• 3. Backbone generation
• 4. Loop modeling
• 5. Side-chain modeling
• 6. Model optimization
• 7. Model validation
• Step 1: Template Recognition and Initial
Alignment
The percentage identity between the sequence
of interest and a possible template is high
enough to be detected with simple sequence
alignment programs such as BLAST.
Step 2: Alignment Correction
• To consider more knowledge during the
alignment, one uses the multiple sequence
alignment (MSA) to derive position specific
scoring matrices, also called profiles.
Determination and Generation of Structurally
Conserved Regions (SCRs)
• There are regions in all proteins belonging to
the same protein family that are nearly
identical in their 3D structures.
• structural units of strongly related proteins,
above all α-helices and β -strands, occupy the
same relative orientations throughout the
whole protein family.
Step 3 Backbone Generation
• Significant differences in protein structures
occur preferably in loop regions, structurally
variable regions (SVRs).
• loop search method, looks for peptide
segments in proteins which meet the
specified geometrical criterion.
• The loops are ranked according to goodness of
fit to the desired structure.
• Using this approach a peptide backbone chain
is built between two conserved segments
using randomly generated numerical values
for all the backbone dihedral angles.
Step 4 Loop Modeling
There are two main approaches to loop
modeling:
1.Knowledge based: one searches the PDB for
known loops with endpoints that match the
residues between which the loop has to be
inserted, and simply copies the loop
conformation.
2. Energy based: an energy function is used to
judge the quality of a loop. Then this function
is minimized, using Monte Carlo (Simons et al.,
1999) or molecular dynamics techniques.
Step 5 Side-Chain Modeling
When we compare the side-chain
conformations (rotamers) of residues that are
conserved in structurally similar proteins, we
find that the often have similar χ1-angles (i.e.,
the torsion angle about the Cα−Cβ bond).
• Used the libraries of common rotamers
extracted from high resolution X-ray
structures.
• The various rotamers are trie successively and
scored with a variety of energy functions.
Step 6: Model Optimization
• Optimizing a complete protein requires an
enormous accuracy in the energy function.
• In short, model optimization does not work
until energy functions (force fields) get more
accurate.
• Two ways to achieve that accuracy are
currently being pursued:
• 1. Quantum force fields
• 2. Self-parameterizing force fields:
• The most straightforward approach to model
optimization is simply to run a molecular
dynamics simulation (MD)of the model. Such a
simulation follows the motions of the protein
on a femtosecond (10−15 s) timescale and
mimics the true folding process.
• The advantage is that a molecular dynamics
simulation implicitly contains entropic effects
that are otherwise difficult to treat.
Step 7 Model Validation
Every homology model contains errors. The
number of errors (for a given method)mainly
depends on two values:
1. The percentage sequence identity between
template and target.
• If it is greater than 90%, the accuracy of the
model can be compared to crystallographically
• determined structures.
• From 50% to 90% identity, the rms error in the
modeled coordinates can be as large as 1.5 A° .
• If the sequence identity drops to 25%, often
leading to very large errors.
2. The number of errors in the template.
There are two principally different ways to
estimate errors in a structure:
Calculating the model’s energy based on a
force field.
Determination of normality indices that
describe how well a given characteristic of the
model resembles the same characteristic in
real structures.
Thank You…..
You might also like
- Homology ModellingDocument21 pagesHomology ModellingJerome FrancisNo ratings yet
- Homology ModelingDocument22 pagesHomology ModelingBasab GhoshNo ratings yet
- Protein ModellingDocument15 pagesProtein ModellingNandini KotharkarNo ratings yet
- Pre-Assessment QuestionsDocument18 pagesPre-Assessment Questionsk.sachinNo ratings yet
- Protein Structure Prediction TechniquesDocument30 pagesProtein Structure Prediction TechniquesJhanvi SNo ratings yet
- 3D Structure PredictionDocument33 pages3D Structure PredictionAyesha KhanNo ratings yet
- Homology Modeling Technique for Protein Structure PredictionDocument19 pagesHomology Modeling Technique for Protein Structure PredictionJainendra JainNo ratings yet
- Homology Modelling: Structure Prediction Using TemplatesDocument19 pagesHomology Modelling: Structure Prediction Using TemplatesManoharNo ratings yet
- Bioinformatics Notes - 17Bt54: Module - 4Document48 pagesBioinformatics Notes - 17Bt54: Module - 4Samuel PrinceNo ratings yet
- Tertiary Structure Prediction Methods: Any Given Protein SequenceDocument29 pagesTertiary Structure Prediction Methods: Any Given Protein Sequencesurrender003No ratings yet
- Homology ModellingDocument29 pagesHomology ModellingRISHITA CHAUHANNo ratings yet
- Protein ModellingDocument53 pagesProtein ModellingiluvgermieNo ratings yet
- Genome Sequencing Projects: Increase in The Number of Protein SequencesDocument27 pagesGenome Sequencing Projects: Increase in The Number of Protein SequencesBiju ThomasNo ratings yet
- Homologymodeling 150123025144 Conversion Gate01Document27 pagesHomologymodeling 150123025144 Conversion Gate01Jhanvi SNo ratings yet
- 3D Structure of Proteins Prediction MethodsDocument9 pages3D Structure of Proteins Prediction MethodsladykikyoyunaNo ratings yet
- Homologymodelingmodeller 181002182751Document19 pagesHomologymodelingmodeller 181002182751Jhanvi SNo ratings yet
- Homology Modelling: A 5-Step ProcessDocument30 pagesHomology Modelling: A 5-Step ProcesskamleshNo ratings yet
- Lesson 2Document17 pagesLesson 2Vamsi Krishna ChaitanyaNo ratings yet
- Re Comb 2003Document16 pagesRe Comb 2003mustafaNo ratings yet
- Bif501 AssDocument3 pagesBif501 Asssana 568aNo ratings yet
- HomolgyDocument21 pagesHomolgySatish SahuNo ratings yet
- Protein Tertiaty Structure PredictionDocument12 pagesProtein Tertiaty Structure PredictionRoshan Mammen ChakkungalNo ratings yet
- Protein Modeling: Protein Structure Prediction Other TopicsDocument76 pagesProtein Modeling: Protein Structure Prediction Other Topicsuma-chenNo ratings yet
- Prediction of Loop Geometries Using A Generalized Born Model of Solvation EffectsDocument11 pagesPrediction of Loop Geometries Using A Generalized Born Model of Solvation EffectsLata DeshmukhNo ratings yet
- Computer-Aided Drug Design (CADD)Document15 pagesComputer-Aided Drug Design (CADD)Amar AlansiNo ratings yet
- Protein Modelling MethodsDocument19 pagesProtein Modelling MethodsrednriNo ratings yet
- Protein ModelingDocument17 pagesProtein ModelingManeeshAgrawalNo ratings yet
- Roy Et Al. - 2011 - A Protocol For Computer-Based Protein Structure AnDocument10 pagesRoy Et Al. - 2011 - A Protocol For Computer-Based Protein Structure Anadiav PakNo ratings yet
- Ab Initio Protein Structure PredictionDocument25 pagesAb Initio Protein Structure PredictionVishnu AjithNo ratings yet
- Molecular Modelling & Drug DesignDocument52 pagesMolecular Modelling & Drug DesignArthe RajarajanNo ratings yet
- Mol - Modelling of BCL-2 FinalDocument37 pagesMol - Modelling of BCL-2 FinalVannakumar SubramanianNo ratings yet
- Structural Biology: What Does 3D Tell Us?Document20 pagesStructural Biology: What Does 3D Tell Us?Prakash KumarNo ratings yet
- Sanchez CurrOpinStructBiol 1997Document9 pagesSanchez CurrOpinStructBiol 1997Phoenix RisingNo ratings yet
- Molecular Dynamics (MD) Simulation in Drug DesignDocument18 pagesMolecular Dynamics (MD) Simulation in Drug DesignSudarshan LamichhaneNo ratings yet
- Unit 3Document9 pagesUnit 3document3580No ratings yet
- Lecture 18Document53 pagesLecture 18IndoBoruto OfficialNo ratings yet
- Shimul SirDocument27 pagesShimul SirTouhid HossainNo ratings yet
- Generative Modeling for Protein StructuresDocument12 pagesGenerative Modeling for Protein Structurescarlos ArozamenaNo ratings yet
- Lecture 101Document43 pagesLecture 101Jean ReneNo ratings yet
- Methods For Accurate Homology Modeling by Global OptimizationDocument17 pagesMethods For Accurate Homology Modeling by Global OptimizationUmmuNo ratings yet
- Improved Prediction of Protein Side-Chain Conformations With SCWRL4Document18 pagesImproved Prediction of Protein Side-Chain Conformations With SCWRL4dunbrackNo ratings yet
- Eswar MethodsMolBiol 2008Document25 pagesEswar MethodsMolBiol 2008huouinkyoumaNo ratings yet
- 1 s2.0 S0006349511022739 MainDocument1 page1 s2.0 S0006349511022739 Mainrully1234No ratings yet
- Identification of Modelling TemplatesDocument7 pagesIdentification of Modelling TemplatesLokesh AdhikariNo ratings yet
- Protein Docking & Folding Methods (40chDocument26 pagesProtein Docking & Folding Methods (40chalone-enolaNo ratings yet
- Docking & DynamicsDocument51 pagesDocking & DynamicsIqbal RadetyoNo ratings yet
- CoMFA Comparitive Molecular Field AnalysisDocument13 pagesCoMFA Comparitive Molecular Field AnalysispinkbutterNo ratings yet
- 1 - Sequence Alignment and VisualizationDocument68 pages1 - Sequence Alignment and VisualizationAlisha100% (1)
- Training ProblemDocument12 pagesTraining ProblemjuanpapoNo ratings yet
- Unit 2.1Document77 pagesUnit 2.1Anny morales AguirreNo ratings yet
- Molecular Dynamics Simulations: Dr.M.Chandra SekharDocument24 pagesMolecular Dynamics Simulations: Dr.M.Chandra SekharchandraloveNo ratings yet
- Sequence Analysis in BioinformaticsDocument18 pagesSequence Analysis in BioinformaticsBhaskar ChatterjeeNo ratings yet
- Lecture 14bDocument46 pagesLecture 14bDishant kumar yadav mhakhariyaNo ratings yet
- Proclust:: Improved Clustering of Protein Sequences With An Extended Graph-Based ApproachDocument58 pagesProclust:: Improved Clustering of Protein Sequences With An Extended Graph-Based ApproachJJ EBNo ratings yet
- Homology PresentationDocument9 pagesHomology PresentationMehwsihNo ratings yet
- Resume 6 The State of Charge Estimating Methods For BatteryDocument28 pagesResume 6 The State of Charge Estimating Methods For BatteryIhsan BayuNo ratings yet
- Text Kuliah Metoda Statistika QSAR KIMED 2023 PDFDocument22 pagesText Kuliah Metoda Statistika QSAR KIMED 2023 PDFameljpNo ratings yet
- ARDL QuestionnaireDocument10 pagesARDL Questionnaire206919No ratings yet
- Protein Modeling by Multiple Sequence Threading and Distance GeometryDocument5 pagesProtein Modeling by Multiple Sequence Threading and Distance GeometryLata DeshmukhNo ratings yet
- Cysteine, Methionine, ProlineDocument3 pagesCysteine, Methionine, ProlineRio BurlazaNo ratings yet
- A Double-Strain TM (gp45) Polypeptide Antigen and Its Application in The Serodiadnosis of Equine Infectius AnemiaDocument8 pagesA Double-Strain TM (gp45) Polypeptide Antigen and Its Application in The Serodiadnosis of Equine Infectius AnemiaFredy MoralesNo ratings yet
- AlphaDocument28 pagesAlphanandugulluNo ratings yet
- Bmi2601 3 2018-Study-Guide PDFDocument124 pagesBmi2601 3 2018-Study-Guide PDFEmeldaNo ratings yet
- LS3 Johnson W13 MT1 Form BDocument9 pagesLS3 Johnson W13 MT1 Form BMarissa ClarkNo ratings yet
- Name: Grade 12 - AMETHYST Date: 2 Summative Exam - Physical Science (Quarter 3)Document2 pagesName: Grade 12 - AMETHYST Date: 2 Summative Exam - Physical Science (Quarter 3)Jeff Tristan CaliganNo ratings yet
- Grade 10 Science Exam Covers DNA, RNA, ProteinsDocument5 pagesGrade 10 Science Exam Covers DNA, RNA, ProteinsNenia Claire Mondarte CruzNo ratings yet
- DNA Codon AlphabetDocument2 pagesDNA Codon AlphabetOmar ElhosaryNo ratings yet
- % Chapter 27: Molecular GeneticsDocument39 pages% Chapter 27: Molecular Geneticsdds uwuNo ratings yet
- 2023 TancetDocument29 pages2023 TancetTechyy SavyNo ratings yet
- Biodegredation of UrethaneDocument8 pagesBiodegredation of UrethaneFuat TopuzNo ratings yet
- Test Bank For Biochemistry 7th Edition Jeremy M BergDocument6 pagesTest Bank For Biochemistry 7th Edition Jeremy M BergkhuyenhenryzwoowNo ratings yet
- JCB 201903033Document20 pagesJCB 201903033Purine ChengNo ratings yet
- IOSR JournalsDocument4 pagesIOSR JournalsInternational Organization of Scientific Research (IOSR)No ratings yet
- Comparative Evaluation of Alternate Emergency FixativesDocument14 pagesComparative Evaluation of Alternate Emergency FixativesIJAR JOURNALNo ratings yet
- 2017 Article 36 PDFDocument10 pages2017 Article 36 PDFmiguelNo ratings yet
- Efolio - Tercenio, Elijah Belle B., BSN 1eDocument60 pagesEfolio - Tercenio, Elijah Belle B., BSN 1e둡챙브로No ratings yet
- Final Exam Study Guide TipsDocument22 pagesFinal Exam Study Guide Tipsrazgriz1211No ratings yet
- Protein Supplements: Which One Should You Consider and Do You Even Need It?Document2 pagesProtein Supplements: Which One Should You Consider and Do You Even Need It?Ravinder VarmaNo ratings yet
- Exam Question So Far Save My Exams MARK SCHEMEDocument11 pagesExam Question So Far Save My Exams MARK SCHEMEArooj AbidNo ratings yet
- Recombinant Protein Purification Handbook PDFDocument244 pagesRecombinant Protein Purification Handbook PDFgicevep758No ratings yet
- Human Nutrition, NotesDocument470 pagesHuman Nutrition, NotesruthNo ratings yet
- Overview of Systems Biology and Omics Technologies: Current Medicinal Chemistry September 2016Document11 pagesOverview of Systems Biology and Omics Technologies: Current Medicinal Chemistry September 2016Janescu LucianNo ratings yet
- ProtibelDocument6 pagesProtibelAna Cláudia AlencarNo ratings yet
- Third Term ss3 ChemistryDocument105 pagesThird Term ss3 ChemistryAdeola OmoniyiNo ratings yet
- Edexcel IGCSE Biology: Structure and Functions Exam QuestionsDocument53 pagesEdexcel IGCSE Biology: Structure and Functions Exam QuestionsZoonieFRNo ratings yet
- Review Methods 1Document12 pagesReview Methods 1João Carloni FilhoNo ratings yet
- SRM University B. Tech Biology for Engineers Course Lesson PlanDocument2 pagesSRM University B. Tech Biology for Engineers Course Lesson Plansanthi saranyaNo ratings yet
- Bakteri RhizosporeDocument7 pagesBakteri RhizosporedanilriosNo ratings yet
- Oschman Trauma Energetics SdarticleDocument14 pagesOschman Trauma Energetics Sdarticlebodyworxs100% (1)